We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Kaitlyn Roberts/Sandbox 2
From Proteopedia
< User:Kaitlyn Roberts(Difference between revisions)
| (51 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
= Human Sterol O-acyltransferase = | = Human Sterol O-acyltransferase = | ||
| - | <StructureSection load='6p2p' size='340' side='right' caption='Human Sterol O-acyltranferase' scene='87/879459/Opening_scene/1'> | + | <StructureSection load='6p2p' size='340' side='right' caption='Human Sterol O-acyltranferase dimer unit' scene='87/879459/Opening_scene/1'> |
| - | == | + | == Functional Overview == |
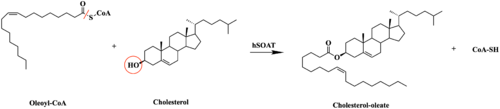
| - | [[Image:SOATfirstreaction.png|500 px|right|thumb|'''Figure 1. | + | [[Image:SOATfirstreaction.png|500 px|right|thumb|'''Figure 1. Esterification reaction of oleoyl-CoA and cholesterol catalyzed by SOAT.''' This reaction results in the formation of cholesteryl esters and CoA-SH byproduct.]] Sterol O-acyltransferase(SOAT), otherwise known as Acyl-coenzyme A:cholesterol acyltransferase(ACAT), is the first discovered member of the membrane-bound O-acyl [https://en.wikipedia.org/wiki/Transferase transferase] or MBOAT enzyme group. MBOAT enzymes are responsible for the transfer of [https://en.wikipedia.org/wiki/Acyl_group acyl chains] onto multiple types of substrates within the cell. There are 11 MBOAT enzyme types that can be found in humans, all of which serve a different function in the overall makeup of human biology.<ref name="Guan">PMID:32424158</ref> |
| - | SOAT specifically catalyzes the [https://en.wikipedia.org/wiki/Fischer–Speier_esterification esterification] of cholesterol for efficient storage within the cell. [https://en.wikipedia.org/wiki/Cholesterol | + | SOAT specifically catalyzes the [https://en.wikipedia.org/wiki/Fischer–Speier_esterification esterification] of cholesterol for efficient storage within the cell (Figure 1). As a membrane lipid, [https://en.wikipedia.org/wiki/Cholesterol cholesterol] is responsible for controlling the fluidity and integrity of the membrane, along with other important biological processes. When there are high concentrations of cholesterol in the cell, [https://en.wikipedia.org/wiki/Cholesteryl_ester cholesteryl esters] can be formed for storage within the membrane.<ref name="Guan" /> |
== Structure == | == Structure == | ||
=== Tertiary Structure === | === Tertiary Structure === | ||
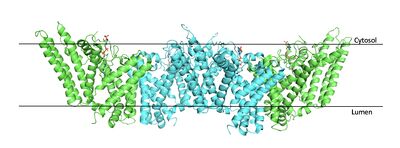
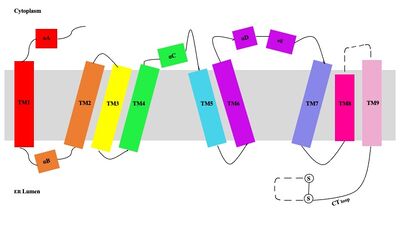
| - | [[Image:Tetramerlabels.jpeg|400 px|right|thumb|'''Figure 2. | + | [[Image:Tetramerlabels.jpeg|400 px|right|thumb|'''Figure 2. Tetramer unit of SOAT shown in position within the membrane.''' The dimer units are identical, as indicated by the corresponding green and blue regions. [http://www.rcsb.org/structure/6P2P PBD 6P2P]]] The biological assembly of SOAT is a <scene name='87/877559/Tetramer_final/2'>tetramer</scene> or a <scene name='87/877559/Dimer_of_dimers_final/2'>dimer of dimers</scene> (Figure 2). Functionally, the <scene name='87/877559/Dimer_final/1'>dimer</scene> units of SOAT are identical and are stabilized by hydrophobic [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] between residues at the <scene name='87/877559/Dimer_interface_final/1'>dimer interface</scene>. Mutating these residues inhibits enzyme activity, suggesting that the dimer unit of SOAT is critical for enzyme function.<ref name="Guan" /> Each dimer consists of two identical <scene name='87/877559/Monomer/7'>monomer</scene> units, individually made up of nine <scene name='87/877559/Transmembrane_helices_final/4'>transmembrane helices</scene> labeled TM1 through TM9 (Figure 3). [[Image:Helicesdiagram1.jpeg|400 px|right|thumb|'''Figure 3. Labeled helices of SOAT within the membrane.''']] The van der Waals interactions at the dimer interface stabilize the dimer between the TM1 helix of one monomer unit and the TM6 [https://en.wikipedia.org/wiki/Lumen_(anatomy) lumenal] segment and TM9 [https://en.wikipedia.org/wiki/Cytosol cytosolic] segment of the other monomer unit. Essential helices (TM1, TM5, TM6, and TM9) from the two monomers form the entrance tunnels and catalytic active site.<ref name="Qian">PMID:32433614</ref> |
=== Tunnel System === | === Tunnel System === | ||
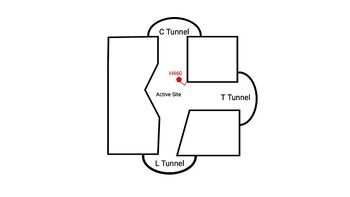
| - | + | An important structural element of SOAT is the tunnel system through which substrates enter and exit. [[Image:2D_Tunnels.jpg|350 px|right|thumb|'''Figure 4. 2D layout of the proposed SOAT tunnel system.''' The C tunnel opens into the cytosol and the L tunnel opens to the lumen. The T tunnel opens into the membrane, but is not quite oriented at a 90 degree angle as depicted here.]] There are three main tunnels in each monomer: the cytosolic (C) tunnel opens to the cytosol, the transmembrane (T) tunnel opens to the membrane, and the lumenal (L) tunnel opens to the lumen (Figure 4). <ref name="Qian" /> The <scene name='87/879459/Ctunnel_final/1'>C tunnel</scene> is the entrance site for the acyl-CoA substrates, allowing them acess to the active site. Residues <scene name='87/877559/C_tunnel_and_measurements_fina/1'>N415, Y433, and K445</scene> exhibit hydrogen bonding interactions with polar atoms of coenzyme A to help stabilize the substrate within the binding pocket. Surface representations of SOAT indicate that there are two alpha helices that block the entrance to the C tunnel, therefore a conformational change needs to occur before the substrate can enter the tunnel. The <scene name='87/877561/Ttunnel_final/1'>T tunnel</scene> opens into the membrane and provides access for cholesterol to enter the active site. At the active site, SOAT catalizes the esterification reaction and the products exit through the tunnels. The CoA-SH product exits through the C tunnel and is released back into the cytosol. The cholesteryl ester product exits through either the T tunnel into the membrane or through the <scene name='87/877561/Ltunnelfinal/1'>L tunnel</scene> | |
| + | into the lumen of the cell. <ref name="Qian" /> | ||
=== Active Site === | === Active Site === | ||
| - | + | Within the binding pocket, there are several <scene name='87/877561/Important_residues_2_final/1'>highly conserved residues</scene>. Their high preservation suggests that the local environment of the binding pocket plays a major role in SOAT activity, but their specific interactions are currently not well studied. However, <scene name='87/877561/Important_residues_final/1'>W420, N421, and H460</scene> have been identified as key catalytic residues.<ref name="Guan" /> Histidine, commonly used as the catalytic base to initiate acyl transferase reactions, is assumed to be the most important catalytic residue in SOAT.<ref name="Das">PMID:18480028</ref> This was confirmed as mutating H460 to alanine completely abolished enzymatic activity.<ref name="Guo">PMID:16154994</ref> Additionally, H460 is highly conserved across a variety of species, further emphasizing its importance in SOAT catalysis.<ref name="Guan" /> It is hypothesized that N421 stabalizes the transition state via hydrogen bonding with coenzyme A.<ref name="Qian" /> Additionally, mutations of W420 to alanine render SOAT nonfunctional, indicating that it must be essential for catalytic activity. However, its role in the mechanism is not explicitly hypothesized. We believe that it plays a role in substrate binding through <scene name='87/879459/W420_intx/4'>hydrophobic interactions</scene> with the acyl chain of coenzyme A. | |
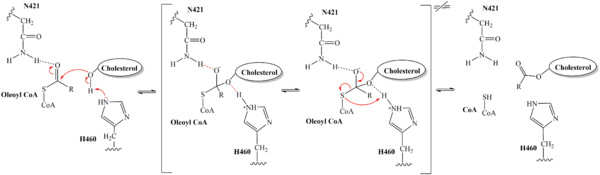
=== Catalytic Mechanism === | === Catalytic Mechanism === | ||
| - | The distal-most nitrogen on H460 acts as a base catalyst to deprotonate the hydroxyl group of | + | The substrate of interest, <scene name='87/879459/Oleoyl-coa_in_bp/2'>oleoyl-CoA</scene>, is shown bound to SOAT to visualize the binding pocket. It must be noted that cholesterol, the other substrate involved, was never correctly imaged in the active site of SOAT. Upon binding of oleoyl-CoA and cholesterol to the SOAT <scene name='87/877555/As_acylcoa_interaction/2'>active site</scene>, the distal-most nitrogen on H460 acts as a base catalyst to deprotonate the hydroxyl group of cholesterol. This leaves the cholesterol oxygen with a negative charge, making it a good [https://en.wikipedia.org/wiki/Nucleophile nucleophile]. The nucleophilic oxygen then attacks oleoyl-CoA at its carbonyl carbon, kicking electron density up to the carbonyl oxygen. The transition state is stabilized by <scene name='87/879459/As_acylcoa_interaction/3'>hydrogen bonding from N421</scene> and the newly protonated H460 (Figure 5). [[Image:6p2pMechanism.png|600 px|right|thumb|'''Figure 5. Mechanism for the esterification reaction of SOAT with arrow pushing.''']] |
| - | From the transition state, excess electron density on the carbonyl oxygen is collapsed back into a double bond. This causes the bond between the carbonyl carbon and sulfur to break, shifting electron density to the sulfur atom. To complete the mechanism, the negatively charged sulfur would reclaim the hydrogen from protonated H460. | + | From the transition state, excess electron density on the carbonyl oxygen is collapsed back into a double bond. This causes the bond between the carbonyl carbon and sulfur to break, shifting electron density to the sulfur atom. To complete the mechanism, the negatively charged sulfur would reclaim the hydrogen from protonated H460 (Figure 5). CoA-SH would exit the active site as a [https://en.wikipedia.org/wiki/Leaving_group leaving group], leaving its oleoyl chain attached to cholesterol in the form of a cholesteryl ester. |
| - | It should be noted that this mechanism is largely hypothesized. Further analysis is needed to confirm the proposed steps | + | It should be noted that this mechanism is largely hypothesized based on our understanding of esterification reactions and the residues involved. Further analysis is needed to confirm the proposed steps. |
== Inhibitors == | == Inhibitors == | ||
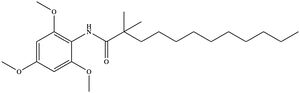
| - | + | [[Image:InhibitorCI976.jpeg|300 px|right|thumb|'''Figure 6. Structure of the CI-976 inhibitor.''']] Depending on concentration and exposure, SOAT activity can be inhibited by CI-976 (Figure 6). When exposed, CI-976 locks itself in the <scene name='87/877559/Inhibitor_in_active_site_final/1'>catalytic center</scene> of the enzyme. The <scene name='87/877559/Residues_and_inhibitor/20'>trimethoxyphenyl head</scene> becomes sandwiched between the catalytic residues W420, N421, and H460. The interaction with these residues, as well as the location of the trimethoxyphenyl head, indicate that CI-976 acts as a [https://en.wikipedia.org/wiki/Competitive_inhibition competitive inhibitor] of SOAT by preventing substrates from accessing the catalytic center. Similar to interactions with the substrates, mutating the key catalytic residues (N421A, H460A, and H460N) results in a smaller effect of the inhibitor on the thermostability of the enzyme.<ref name="Guan" /> | |
== Biological Relevance == | == Biological Relevance == | ||
| - | SOAT | + | SOAT has gained interest when looking at its biological relevance because it has the ability to use a wide range of sterols in it’s mechanistic activity. The wide variety of substrates has led to SOAT being focused on as a potential drug target for many different diseases. [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer’s disease], [https://en.wikipedia.org/wiki/Atherosclerosis atherosclerosis], and several types of cancers have show success in treatments when targeting the SOAT enzyme’s catalytic mechanism. <ref name="Guan" /> In summary, targeting SOAT could be an effective means for treating various diseases. Aberrant quantities of cholesteryl esters seem to hinder various cellular processes; thus, inhibiting SOAT expression and functionality could help reduce these adverse effects. Overall, SOAT plays an important role in cholesterol homeostasis and future research of this enzyme could lead to the discovery of therapeutic treatments for different illnesses. |
| + | |||
| + | ==== Alzheimer's Disease ==== | ||
Alzheimer’s disease (AD) is a progressive disease that severely hinders a person’s memory and other cognitive functions. AD is the result of a significant increase in [https://en.wikipedia.org/wiki/Amyloid_beta beta-amyloid] (Aβ) peptide concentration. <ref name="Bhattacharyya">PMID:20398792</ref> Previous studies have found that the amount and distribution of intracellular cholesterol plays an important role in regulating Aβ production.<ref name="Huttunen">PMID:18322393</ref> Therefore, SOAT inhibition could be an effective therapy for treating AD because it would reduce cholesteryl ester formation in the brain and help lower Aβ generation as well. <ref name="Bhattacharyya" /> <ref name="Huttunen" /> | Alzheimer’s disease (AD) is a progressive disease that severely hinders a person’s memory and other cognitive functions. AD is the result of a significant increase in [https://en.wikipedia.org/wiki/Amyloid_beta beta-amyloid] (Aβ) peptide concentration. <ref name="Bhattacharyya">PMID:20398792</ref> Previous studies have found that the amount and distribution of intracellular cholesterol plays an important role in regulating Aβ production.<ref name="Huttunen">PMID:18322393</ref> Therefore, SOAT inhibition could be an effective therapy for treating AD because it would reduce cholesteryl ester formation in the brain and help lower Aβ generation as well. <ref name="Bhattacharyya" /> <ref name="Huttunen" /> | ||
| + | |||
| + | ==== Atherosclerosis ==== | ||
Another disease SOAT inhibition could help treat is atherosclerosis. Buildup of cholesteryl esters from SOAT catalysis has been shown to be partially responsible for foam cell formation, one of the major indicators of atherosclerosis. Consequently, SOAT inhibitors have been studied as potential drug targets for this disease.<ref name="Chang">PMID:16518538</ref> | Another disease SOAT inhibition could help treat is atherosclerosis. Buildup of cholesteryl esters from SOAT catalysis has been shown to be partially responsible for foam cell formation, one of the major indicators of atherosclerosis. Consequently, SOAT inhibitors have been studied as potential drug targets for this disease.<ref name="Chang">PMID:16518538</ref> | ||
| - | Increased expression of SOAT and abnormal accumulation of cholesteryl esters has also been found in multiple cancers including ovarian cancer. Therefore, inhibiting SOAT and exhausting cholesteryl ester concentrations has shown to have anti-tumor effects in terms of monitoring apoptosis, cell proliferation, and migration and invasion properties. Therapies that target SOAT regulation and expression levels could thus lead to potential treatments for ovarian and other types of cancer.<ref name="Ayyagari">PMID:31978092</ref> | + | |
| - | + | ==== Cancer ==== | |
| - | + | Increased expression of SOAT and abnormal accumulation of cholesteryl esters has also been found in multiple cancers including [https://en.wikipedia.org/wiki/Ovarian_cancer ovarian cancer]. Therefore, inhibiting SOAT and exhausting cholesteryl ester concentrations has shown to have anti-tumor effects in terms of monitoring [https://en.wikipedia.org/wiki/Apoptosis apoptosis], [https://en.wikipedia.org/wiki/Cell_proliferation cell proliferation], and migration and invasion properties. Therapies that target SOAT regulation and expression levels could thus lead to potential treatments for ovarian and other types of cancer.<ref name="Ayyagari">PMID:31978092</ref> | |
| + | |||
</StructureSection> | </StructureSection> | ||
Current revision
Human Sterol O-acyltransferase
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ 2.0 2.1 2.2 2.3 Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Das A, Davis MA, Rudel LL. Identification of putative active site residues of ACAT enzymes. J Lipid Res. 2008 Aug;49(8):1770-81. doi: 10.1194/jlr.M800131-JLR200. Epub 2008, May 13. PMID:18480028 doi:http://dx.doi.org/10.1194/jlr.M800131-JLR200
- ↑ Guo ZY, Lin S, Heinen JA, Chang CC, Chang TY. The active site His-460 of human acyl-coenzyme A:cholesterol acyltransferase 1 resides in a hitherto undisclosed transmembrane domain. J Biol Chem. 2005 Nov 11;280(45):37814-26. doi: 10.1074/jbc.M508384200. Epub 2005, Sep 8. PMID:16154994 doi:http://dx.doi.org/10.1074/jbc.M508384200
- ↑ 5.0 5.1 Bhattacharyya R, Kovacs DM. ACAT inhibition and amyloid beta reduction. Biochim Biophys Acta. 2010 Aug;1801(8):960-5. doi: 10.1016/j.bbalip.2010.04.003. , Epub 2010 Apr 14. PMID:20398792 doi:http://dx.doi.org/10.1016/j.bbalip.2010.04.003
- ↑ 6.0 6.1 Huttunen HJ, Kovacs DM. ACAT as a drug target for Alzheimer's disease. Neurodegener Dis. 2008;5(3-4):212-4. doi: 10.1159/000113705. Epub 2008 Mar 6. PMID:18322393 doi:http://dx.doi.org/10.1159/000113705
- ↑ Chang C, Dong R, Miyazaki A, Sakashita N, Zhang Y, Liu J, Guo M, Li BL, Chang TY. Human acyl-CoA:cholesterol acyltransferase (ACAT) and its potential as a target for pharmaceutical intervention against atherosclerosis. Acta Biochim Biophys Sin (Shanghai). 2006 Mar;38(3):151-6. doi:, 10.1111/j.1745-7270.2006.00154.x. PMID:16518538 doi:http://dx.doi.org/10.1111/j.1745-7270.2006.00154.x
- ↑ Ayyagari VN, Wang X, Diaz-Sylvester PL, Groesch K, Brard L. Assessment of acyl-CoA cholesterol acyltransferase (ACAT-1) role in ovarian cancer progression-An in vitro study. PLoS One. 2020 Jan 24;15(1):e0228024. doi: 10.1371/journal.pone.0228024., eCollection 2020. PMID:31978092 doi:http://dx.doi.org/10.1371/journal.pone.0228024
Student Contributors
- Kylie Pfeifer
- Stephanie Pellegrino
- Kaitlyn Roberts