Lysine-specific demethylase 1 (LSD-1)
From Proteopedia
(Difference between revisions)
(Category:Featured in BAMBED. Date must be of the page, not the paper) |
|||
| (5 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | {{BAMBED | ||
| + | |DATE=June 14, 2021 | ||
| + | |OLDID=3412175 | ||
| + | |BAMBEDDOI=10.1002/bmb.21759 | ||
| + | }} | ||
=Human lysine-specific demethylase 1 (LSD-1), A repressor of transcription= | =Human lysine-specific demethylase 1 (LSD-1), A repressor of transcription= | ||
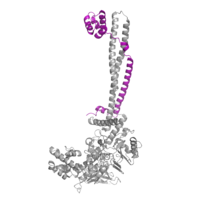
<StructureSection load='2h94' size='330' frame='true' side='right' caption='LSD-1 (PDB: [[2h94]]) overall 3D structure: Tower domain (blue), SWIRM domain (yellow), Oxidase domain (orange), and FAD cofactor (green).' scene='83/834203/Overall_lsd-1/1'> | <StructureSection load='2h94' size='330' frame='true' side='right' caption='LSD-1 (PDB: [[2h94]]) overall 3D structure: Tower domain (blue), SWIRM domain (yellow), Oxidase domain (orange), and FAD cofactor (green).' scene='83/834203/Overall_lsd-1/1'> | ||
| Line 4: | Line 9: | ||
== Introduction == | == Introduction == | ||
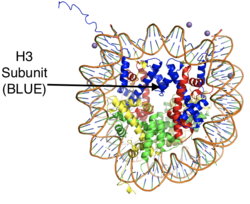
[[Image:Histonemm.png |250px|right|thumb| Figure 1: The crystal structure of a histone core octamer (each subunit in a different color) bound to DNA. PDB:5y0c]] | [[Image:Histonemm.png |250px|right|thumb| Figure 1: The crystal structure of a histone core octamer (each subunit in a different color) bound to DNA. PDB:5y0c]] | ||
| - | <scene name='83/834203/Overall_lsd-1/1'>LSD-1</scene>, '''human lysine-specific demethylase 1,''' is an enzyme that affects the ability of DNA to associate with [https://proteopedia.org/wiki/index.php/Nucleosome histone proteins]. Histone proteins contain residues that give them an overall positive charge and allow them to act as spools upon which negatively charged DNA can wrap around for storage in the nucleus of a cell (Figure 1). When DNA is tightly condensed it forms into [https://proteopedia.org/wiki/index.php/Nucleosome_structure nucleosomes] which consist of eight histone core proteins (2 H2A, 2 H2B, 2 H3, 2 H4) with DNA tightly coiled around them. This tightly coiled DNA is known as [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin], which is inaccessible to [https://en.wikipedia.org/wiki/Transcription_factor transcription factors] and [https://en.wikipedia.org/wiki/RNA_polymerase RNA polymerase]. This can be reversed by [https://en.wikipedia.org/wiki/Post-translational_modification modifications] to the histone protein structure that cause the DNA to relax and form [https://en.wikipedia.org/wiki/Euchromatin euchromatin], which allows for gene expression. One key histone modification is the [https://en.wikipedia.org/wiki/Methyltransferase methylation] and subsequent [https://en.wikipedia.org/wiki/Demethylase demethylation] of lysine residues. Before 2004, it was believed that methylation of histone tails was stable and irreversible. In 2004, it was discovered that histone tails can also be demethylated by demethylase enzymes such as LSD-1.<ref name="Shi">doi: 10.1016/j.cell.2004.12.012</ref> | + | <scene name='83/834203/Overall_lsd-1/1'>LSD-1</scene>, '''human lysine-specific demethylase 1,''' is an enzyme that affects the ability of DNA to associate with [https://proteopedia.org/wiki/index.php/Nucleosome histone proteins]. Histone proteins contain residues that give them an overall positive charge and allow them to act as spools upon which negatively charged DNA can wrap around for storage in the nucleus of a cell (Figure 1). When DNA is tightly condensed it forms into [https://proteopedia.org/wiki/index.php/Nucleosome_structure nucleosomes] which consist of eight histone core proteins (2 H2A, 2 H2B, 2 H3, 2 H4) with DNA tightly coiled around them. This tightly coiled DNA is known as [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin], which is inaccessible to [https://en.wikipedia.org/wiki/Transcription_factor transcription factors] and [https://en.wikipedia.org/wiki/RNA_polymerase RNA polymerase]. This can be reversed by [https://en.wikipedia.org/wiki/Post-translational_modification post-translational modifications] to the histone protein structure that cause the DNA to relax and form [https://en.wikipedia.org/wiki/Euchromatin euchromatin], which allows for gene expression. One key histone modification is the [https://en.wikipedia.org/wiki/Methyltransferase methylation] and subsequent [https://en.wikipedia.org/wiki/Demethylase demethylation] of lysine residues. Before 2004, it was believed that methylation of histone tails was stable and irreversible. In 2004, it was discovered that histone tails can also be demethylated by demethylase enzymes such as LSD-1.<ref name="Shi">doi: 10.1016/j.cell.2004.12.012</ref> |
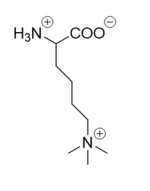
There are two main classes of demethylases, and they are categorized by their cofactors and reaction mechanisms. The KDM1 family of demethylases employs a flavin adenine dinucleotide [https://en.wikipedia.org/wiki/Flavin_adenine_dinucleotide (FAD)] cofactor to catalyze the demethylation reaction. The other class of demethylases consists of five families (KDM2-6) and use an Fe<sup>2+</sup> ion cofactor and [https://en.wikipedia.org/wiki/Alpha-Ketoglutaric_acid α-ketoglutarate] as a cosubstrate to catalyze the reaction. Although the cofactors used are different, both classes operate by hydroxylating the target methyl group. LSD-1 is in the KDM1 family of histone demethylases that uses FAD as a cofactor. LSD-1 specifically demethylates mono- or di-methylated lysine substrates at Lys4 or Lys9 in the tail of histone H3.<ref name="Forneris">PMID: 15811342</ref> Demethylation of these lysine residues is commonly associated with transcriptional activation, but it also has the ability to silence genes depending on the residue being demethylated, the cofactors present, and the environment in which the demethylation occurs. | There are two main classes of demethylases, and they are categorized by their cofactors and reaction mechanisms. The KDM1 family of demethylases employs a flavin adenine dinucleotide [https://en.wikipedia.org/wiki/Flavin_adenine_dinucleotide (FAD)] cofactor to catalyze the demethylation reaction. The other class of demethylases consists of five families (KDM2-6) and use an Fe<sup>2+</sup> ion cofactor and [https://en.wikipedia.org/wiki/Alpha-Ketoglutaric_acid α-ketoglutarate] as a cosubstrate to catalyze the reaction. Although the cofactors used are different, both classes operate by hydroxylating the target methyl group. LSD-1 is in the KDM1 family of histone demethylases that uses FAD as a cofactor. LSD-1 specifically demethylates mono- or di-methylated lysine substrates at Lys4 or Lys9 in the tail of histone H3.<ref name="Forneris">PMID: 15811342</ref> Demethylation of these lysine residues is commonly associated with transcriptional activation, but it also has the ability to silence genes depending on the residue being demethylated, the cofactors present, and the environment in which the demethylation occurs. | ||
| Line 19: | Line 24: | ||
=== Oxidase Domain === | === Oxidase Domain === | ||
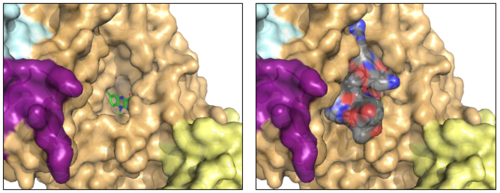
| - | + | The <scene name='83/834203/Oxidasedomain/4'>oxidase domain</scene> houses the catalytic site of LSD-1. This domain non-covalently binds the FAD cofactor and the substrate lysine on the H3 histone tail.<ref name="Stavropolous"/> The FAD binding cavity is quite large (15 Å deep and around 25 Å wide) in relation to other oxidases that utilize FAD as a cofactor (Figure 3, left panel).<ref name="Stavropolous"/> In comparison, [https://en.wikipedia.org/wiki/Polyamine_oxidase polyamine oxidase], another FAD-dependent oxidase, has a catalytic chamber roughly 30 Å long but only a few angstroms wide.<ref name=”Binda”>PMID:11258887</ref> The relatively large size of the LSD-1 active site cavity is to accommodate the first 15 residues of the histone H3 substrate (Figure 3, right panel). | |
| + | [[Image:lsd_h3_final.png|500px|center|thumb| | ||
| + | Figure 3: The FAD binding cavity in the oxidase domain of LSD-1 (left) and in the presence of histone H3-peptide (right). The swirm domain is yellow, CoREST is purple, the oxidase domain is orange, and the tower domain is light blue. The FAD is shown as green sticks and the H3-peptide is gray. PDB: 2V1D]] | ||
====FAD Cofactor==== | ====FAD Cofactor==== | ||
| Line 30: | Line 37: | ||
== Enzymatic Mechanism == | == Enzymatic Mechanism == | ||
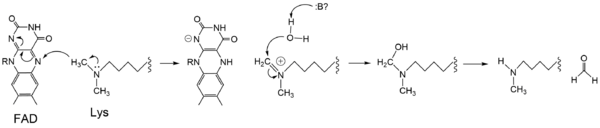
The mechanism of lysine demethylation catalyzed by LSD-1 employs FAD as an oxidizing agent to oxidize the methyl carbon of the lysine substrate (Figure 4). The FAD cofactor, positioned close to the substrate lysine in the active site, initiates a two-electron transfer in the form of a [https://en.wikipedia.org/wiki/Hydride hydride] between the substrate methyl-lysine and the isoallozaxine ring of the FAD. The cofactor becomes anionic and the resonance form is stabilized by the positively charged <scene name='83/834203/Lys661/3'>Lys661</scene> positioned in the catalytic pocket of the active site. The lysine subatrate forms an [https://en.wikipedia.org/wiki/Iminium iminium] cation that is hydrolyzed into the [https://en.wikipedia.org/wiki/Hemiaminal hemiaminal] intermediate, potentially using water and general base catalysis of a nearby residue. The hemiaminal intermediate readily decomposes to formaldehyde and the demethylated lysine.<ref name="Stavropolous"/> | The mechanism of lysine demethylation catalyzed by LSD-1 employs FAD as an oxidizing agent to oxidize the methyl carbon of the lysine substrate (Figure 4). The FAD cofactor, positioned close to the substrate lysine in the active site, initiates a two-electron transfer in the form of a [https://en.wikipedia.org/wiki/Hydride hydride] between the substrate methyl-lysine and the isoallozaxine ring of the FAD. The cofactor becomes anionic and the resonance form is stabilized by the positively charged <scene name='83/834203/Lys661/3'>Lys661</scene> positioned in the catalytic pocket of the active site. The lysine subatrate forms an [https://en.wikipedia.org/wiki/Iminium iminium] cation that is hydrolyzed into the [https://en.wikipedia.org/wiki/Hemiaminal hemiaminal] intermediate, potentially using water and general base catalysis of a nearby residue. The hemiaminal intermediate readily decomposes to formaldehyde and the demethylated lysine.<ref name="Stavropolous"/> | ||
| - | [[Image:LSD_1_Chemdraw_Mech.png| | + | [[Image:LSD_1_Chemdraw_Mech.png|600px|center|thumb|Figure 4: Hydride transfer mechanism catalyzed by LSD-1 with a dimethyl-lysine substrate.]] |
===Inhibition by Tri-Methylated Lysine=== | ===Inhibition by Tri-Methylated Lysine=== | ||
| Line 51: | Line 58: | ||
== Student Contributors == | == Student Contributors == | ||
Nicholas Bantz, Sean Callahan, Cody Carley, Andrew Hesterhagen, Steve Klimcak, Michael Thomas | Nicholas Bantz, Sean Callahan, Cody Carley, Andrew Hesterhagen, Steve Klimcak, Michael Thomas | ||
| + | |||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
| + | [[Category:Featured in BAMBED]] | ||
Current revision
This page, as it appeared on June 14, 2021, was featured in this article in the journal Biochemistry and Molecular Biology Education.
Human lysine-specific demethylase 1 (LSD-1), A repressor of transcription
| |||||||||||
Student Contributors
Nicholas Bantz, Sean Callahan, Cody Carley, Andrew Hesterhagen, Steve Klimcak, Michael Thomas
Proteopedia Page Contributors and Editors (what is this?)
Mark Macbeth, Valentine J Klimkowski, Michal Harel, Angel Herraez