Transcription-repair coupling factor
From Proteopedia
(→Structure and conformational change) |
|||
| (6 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | __TOC__ | ||
The bacterial [[transcription-repair coupling factor]] TRCF, also called Mfd translocase, is a DNA repair protein. It has a role in transcription-coupled repair, a subpathway of nucleotide excision repair (NER). Mfd recognizes stalled RNA polymerase (RNAP) and either restarts transcription or removes the stalled polymerase and recruits the NER proteins UvrA and UvrB. | The bacterial [[transcription-repair coupling factor]] TRCF, also called Mfd translocase, is a DNA repair protein. It has a role in transcription-coupled repair, a subpathway of nucleotide excision repair (NER). Mfd recognizes stalled RNA polymerase (RNAP) and either restarts transcription or removes the stalled polymerase and recruits the NER proteins UvrA and UvrB. | ||
| Line 9: | Line 10: | ||
== Structure and conformational change == | == Structure and conformational change == | ||
<StructureSection load='' size='350' side='right' scene='2eyq/Domainscolorsflabe/3' caption=''> | <StructureSection load='' size='350' side='right' scene='2eyq/Domainscolorsflabe/3' caption=''> | ||
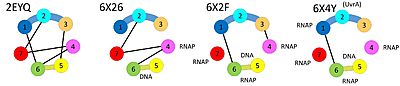
| - | The initial scene (switch to <scene name='46/460252/Apo/1'>cartoon</scene>) shows the domains of Mfd in the conformation of the apo-enzyme.<ref>DOI:10.1016/j.cell.2005.11.045</ref> The UvrA interaction site (on domain 2) is occluded by domain 7. The translocase domains (domain 5 and domain 6), through interactions with domains 1 and 3, are locked in an inactive conformation, preventing the typical hinge motion of translocases when they bind and hydrolyze ATP while moving along DNA. When Mfd binds to DNA in the presence of | + | The initial scene (switch to <scene name='46/460252/Apo/1'>cartoon</scene>) shows the domains of Mfd in the conformation of the apo-enzyme.<ref>DOI:10.1016/j.cell.2005.11.045</ref> The UvrA interaction site (on domain 2) is occluded by domain 7. The translocase domains (domain 5 and domain 6), through interactions with domains 1 and 3, are locked in an inactive conformation, preventing the typical hinge motion of translocases when they bind and hydrolyze ATP while moving along DNA. When Mfd binds to DNA in the presence of ADP·AlFₓ, Mfd undergoes large domain rearrangements, giving a hint how it might bind to stalled RNAP.<ref>DOI:10.1038/s41467-020-17457-1</ref> A first glimpse of the Mfd RNAP complex was through cryo-EM, but large parts of the Mfd protein were not resolved.<ref>DOI:10.1093/nar/gkaa904</ref> |
[[Image:Mfd domains.JPG|thumb|left]] | [[Image:Mfd domains.JPG|thumb|left]] | ||
| - | In 2021, the structures of several intermediates in the presences of stalled RNAP were determined using cryo-EM, shedding light on the loading and activation mechanism, as well as how translocation of Mfd on DNA leads to disruption of the RNAP elongation complex and recruitment of UvrA.<ref>doi: 10.7554/eLife.62117</ref> | + | In 2021, the structures of several intermediates in the presences of stalled RNAP were determined using cryo-EM, shedding further light on the loading and activation mechanism, as well as how translocation of Mfd on DNA leads to disruption of the RNAP elongation complex and recruitment of UvrA.<ref>doi: 10.7554/eLife.62117</ref> |
When <scene name='46/460252/Mfd_rnap_l1/2'>Mfd binds to a stalled RNA polymerase</scene>, the RID (domain 4) binds to the beta subunit of RNA polymerase and domains 5 and domain 6 bind to DNA and ATP. These multiple binding events require a different relative orientation of RID and the translocase domains, and the necessary conformational changes disrupt inter-domain interactions seen in the apo-structure while activating the translocase activity of Mfd. | When <scene name='46/460252/Mfd_rnap_l1/2'>Mfd binds to a stalled RNA polymerase</scene>, the RID (domain 4) binds to the beta subunit of RNA polymerase and domains 5 and domain 6 bind to DNA and ATP. These multiple binding events require a different relative orientation of RID and the translocase domains, and the necessary conformational changes disrupt inter-domain interactions seen in the apo-structure while activating the translocase activity of Mfd. | ||
| Line 25: | Line 26: | ||
In the apo-structure, domains 3 and 4 are far apart from each other (connected by an extended linker), but they make direct contacts when Mfd is loaded on DNA. On the other hand, domains 4 and 5 make direct contacts in the apo-structure, but are far apart from each other (coonected by an extended linker) when Mfd is loaded on DNA. Finally, domain 2 and domain 7 bind to each other in the apo-state whereas domain 2 is available for binding UvrA in the DNA-bound state. | In the apo-structure, domains 3 and 4 are far apart from each other (connected by an extended linker), but they make direct contacts when Mfd is loaded on DNA. On the other hand, domains 4 and 5 make direct contacts in the apo-structure, but are far apart from each other (coonected by an extended linker) when Mfd is loaded on DNA. Finally, domain 2 and domain 7 bind to each other in the apo-state whereas domain 2 is available for binding UvrA in the DNA-bound state. | ||
| - | + | == 3D Structures of Transcription-repair coupling factor == | |
| + | [[Transcription-repair coupling factor 3D structures]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
| - | |||
| - | == 3D Structures of Transcription-repair coupling factor == | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | |||
| - | [[2eyq]] – EcTRCF – ''Escherichia coli''<br /> | ||
| - | [[6x2n]], [[6x2f]], [[6x26]], [[6x50]], [[6x43]], [[6x4w]], [[6x4y]] - EcTRCF, RNA polymerase, RNA, DNA (Cryo EM)<br /> | ||
| - | [[3hjh]] – EcTRCF residues 1-470<br /> | ||
| - | [[2b2n]] - EcTRCF residues 1-333<br /> | ||
| - | [[6yhz]] - EcTRCF residues 472-547 – NMR<br /> | ||
| - | [[4dfc]] – EcTRCF D2 domain 127-213 + UvrABC system protein A<br /> | ||
| - | [[6xeo]] – EcTRCF + DNA – Cryo EM<br /> | ||
| - | [[3mlq]] – TtTRCF RNA polymerase interacting domain + DNA-directed RNA polymerase subunit β - ''Thermus thermophilus''<br /> | ||
| - | [[6m6a]] – TtTRCF + RNA polymerase – Cryo EM<br /> | ||
| - | [[6m6b]] – TtTRCF + RNA polymerase + ATP-γ-S – Cryo EM<br /> | ||
| - | [[2qsr]] – TRCF C terminal – ''Streptococcus pneumoni''<br /> | ||
| - | [[6ac6]], [[6aca]], [[6ac8]] – MsTRCF – ''Mycobacterium smegmatis''<br /> | ||
| - | [[6acx]] – MsTRCF + ADP<br /> | ||
| - | |||
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
| Line 65: | Line 48: | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
<br /> | <br /> | ||
| + | </StructureSection> | ||
Created with the participation of [[User:Wayne Decatur|Wayne Decatur]] | Created with the participation of [[User:Wayne Decatur|Wayne Decatur]] | ||
Current revision
Contents |
The bacterial transcription-repair coupling factor TRCF, also called Mfd translocase, is a DNA repair protein. It has a role in transcription-coupled repair, a subpathway of nucleotide excision repair (NER). Mfd recognizes stalled RNA polymerase (RNAP) and either restarts transcription or removes the stalled polymerase and recruits the NER proteins UvrA and UvrB.
Function
Mfd has ATP hydrolysis activity, DNA binding sites and a UvrA binding sites. These three functions are inhibited in the isolated enzyme, but are activated when Mfd encounters stalled RNA polymerase (or through various mutations that remove inhibitory domains [1]). Mfd also contains an RNA interaction domain (RID) that binds to the beta subunit of RNAP.
Relationship to other enzymes
The N-terminal part of Mfd shows sequence similarity to UvrB, including in the domain of UvrB that interacts with UvrA. However, the conserved helicase motifs present in UvrB (responsible for binding and hydrolyzing ATP) are absent in that part of Mfd. Furthermore, the sequence segment known to fold as a beta hairpin in UvrB (involved in clamping down a single strand of DNA) seems absent. The C-terminal part of Mfd shows sequence similarity to SF1/SF2 helicases (UvrB is an example), containing conserved helicase/translocase motifs.
Structure and conformational change
| |||||||||||
Created with the participation of Wayne Decatur
Proteopedia Page Contributors and Editors (what is this?)
Karsten Theis, Michal Harel, Alexander Berchansky, Wayne Decatur