|
|
| Line 3: |
Line 3: |
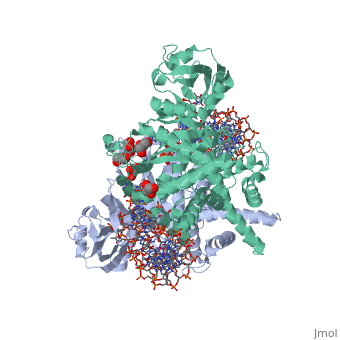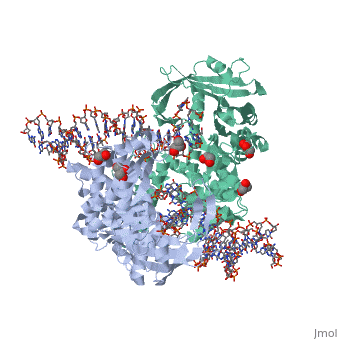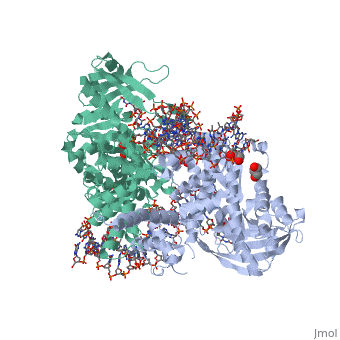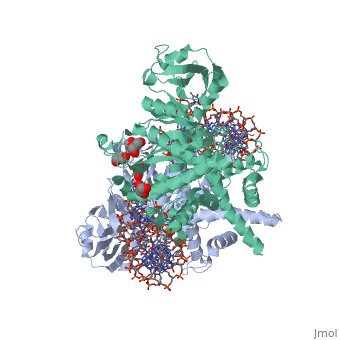
| | <StructureSection load='3ecp' size='340' side='right'caption='[[3ecp]], [[Resolution|resolution]] 2.50Å' scene=''> | | <StructureSection load='3ecp' size='340' side='right'caption='[[3ecp]], [[Resolution|resolution]] 2.50Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[3ecp]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/"bacillus_coli"_migula_1895 "bacillus coli" migula 1895]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3ECP OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3ECP FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[3ecp]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3ECP OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3ECP FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GOL:GLYCEROL'>GOL</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> |
| - | <tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat"><div style='overflow: auto; max-height: 3em;'>[[1muh|1muh]], [[1mur|1mur]], [[1mus|1mus]]</div></td></tr> | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GOL:GLYCEROL'>GOL</scene></td></tr> |
| - | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">orf163 ([https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=562 "Bacillus coli" Migula 1895])</td></tr>
| + | |
| | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3ecp FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3ecp OCA], [https://pdbe.org/3ecp PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3ecp RCSB], [https://www.ebi.ac.uk/pdbsum/3ecp PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3ecp ProSAT]</span></td></tr> | | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3ecp FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3ecp OCA], [https://pdbe.org/3ecp PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3ecp RCSB], [https://www.ebi.ac.uk/pdbsum/3ecp PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3ecp ProSAT]</span></td></tr> |
| | </table> | | </table> |
| | == Function == | | == Function == |
| - | [[https://www.uniprot.org/uniprot/TN5P_ECOLX TN5P_ECOLX]] Mediates transposition of transposon Tn5 by a 'cut and paste' mechanism. First, the monomeric transposase binds the 19 bp inverted DNA repeats flanking the transposon. Then, dimerization of the DNA-bound transposase creates a synaptic DNA complex. After nicking of the first DNA strand, excision of the transposon proceeds through a series of intermediates. The transposase then mediates the insertion of the transposon at a new site by strand transfer. The activity of the wild-type transposase is very low, and is further inhibited by dimerization with the transposase inhibitor (inh).<ref>PMID:6260374</ref> <ref>PMID:6291786</ref> <ref>PMID:6303899</ref> <ref>PMID:1310499</ref> <ref>PMID:8226636</ref> <ref>PMID:8871560</ref> <ref>PMID:11877443</ref> <ref>PMID:12367522</ref>
| + | [https://www.uniprot.org/uniprot/TN5P_ECOLX TN5P_ECOLX] Mediates transposition of transposon Tn5 by a 'cut and paste' mechanism. First, the monomeric transposase binds the 19 bp inverted DNA repeats flanking the transposon. Then, dimerization of the DNA-bound transposase creates a synaptic DNA complex. After nicking of the first DNA strand, excision of the transposon proceeds through a series of intermediates. The transposase then mediates the insertion of the transposon at a new site by strand transfer. The activity of the wild-type transposase is very low, and is further inhibited by dimerization with the transposase inhibitor (inh).<ref>PMID:6260374</ref> <ref>PMID:6291786</ref> <ref>PMID:6303899</ref> <ref>PMID:1310499</ref> <ref>PMID:8226636</ref> <ref>PMID:8871560</ref> <ref>PMID:11877443</ref> <ref>PMID:12367522</ref> |
| | == Evolutionary Conservation == | | == Evolutionary Conservation == |
| | [[Image:Consurf_key_small.gif|200px|right]] | | [[Image:Consurf_key_small.gif|200px|right]] |
| Line 38: |
Line 37: |
| | __TOC__ | | __TOC__ |
| | </StructureSection> | | </StructureSection> |
| - | [[Category: Bacillus coli migula 1895]] | + | [[Category: Escherichia coli]] |
| | [[Category: Large Structures]] | | [[Category: Large Structures]] |
| - | [[Category: Klenchin, V A]] | + | [[Category: Klenchin VA]] |
| - | [[Category: Dna recombination-dna complex]]
| + | |
| - | [[Category: Protein-dna complex]]
| + | |
| - | [[Category: Ribonuclease h-like motif]]
| + | |
| - | [[Category: Synaptic complex]]
| + | |
| - | [[Category: Transposase]]
| + | |
| Structural highlights
Function
TN5P_ECOLX Mediates transposition of transposon Tn5 by a 'cut and paste' mechanism. First, the monomeric transposase binds the 19 bp inverted DNA repeats flanking the transposon. Then, dimerization of the DNA-bound transposase creates a synaptic DNA complex. After nicking of the first DNA strand, excision of the transposon proceeds through a series of intermediates. The transposase then mediates the insertion of the transposon at a new site by strand transfer. The activity of the wild-type transposase is very low, and is further inhibited by dimerization with the transposase inhibitor (inh).[1] [2] [3] [4] [5] [6] [7] [8]
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
Bacterial DNA transposition is an important model system for studying DNA recombination events such as HIV-1 DNA integration and RAG-1-mediated V(D)J recombination. This communication focuses on the role of protein-phosphate contacts in manipulating DNA structure as a requirement for transposition catalysis. In particular, the participation of the nontransferred strand (NTS) 5' phosphate in Tn5 transposition strand transfer is analyzed. The 5' phosphate plays no direct catalytic role, nonetheless its presence stimulates strand transfer approximately 30-fold. X-ray crystallography indicates that transposase-DNA complexes formed with NTS 5' phosphorylated DNA have two properties that contrast with structures formed with complexes lacking the 5' phosphate or complexes generated from in-crystal hairpin cleavage. Transposase residues R210, Y319 and R322 of the (R)YREK motif coordinate the 5' phosphate rather than the subterminal NTS phosphate, and the 5' NTS end is moved away from the 3' transferred strand end. Mutation R210A impairs the 5' phosphate stimulation. It is posited that DNA phosphate coordination by R210, Y319 and R322 results in movement of the 5' NTS DNA away from the 3'-end thus allowing efficient target DNA binding. It is likely that this role for the newly identified RYR triad is utilized by other transposase-related proteins.
Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif.,Klenchin VA, Czyz A, Goryshin IY, Gradman R, Lovell S, Rayment I, Reznikoff WS Nucleic Acids Res. 2008 Oct;36(18):5855-62. Epub 2008 Sep 12. PMID:18790806[9]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Rothstein SJ, Reznikoff WS. The functional differences in the inverted repeats of Tn5 are caused by a single base pair nonhomology. Cell. 1981 Jan;23(1):191-9. PMID:6260374
- ↑ Johnson RC, Yin JC, Reznikoff WS. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982 Oct;30(3):873-82. PMID:6291786
- ↑ Lowe JB, Berg DE. A product of the TN5 transposase gene inhibits transposition. Genetics. 1983 Apr;103(4):605-15. PMID:6303899
- ↑ Wiegand TW, Reznikoff WS. Characterization of two hypertransposing Tn5 mutants. J Bacteriol. 1992 Feb;174(4):1229-39. PMID:1310499
- ↑ de la Cruz NB, Weinreich MD, Wiegand TW, Krebs MP, Reznikoff WS. Characterization of the Tn5 transposase and inhibitor proteins: a model for the inhibition of transposition. J Bacteriol. 1993 Nov;175(21):6932-8. PMID:8226636
- ↑ York D, Reznikoff WS. Purification and biochemical analyses of a monomeric form of Tn5 transposase. Nucleic Acids Res. 1996 Oct 1;24(19):3790-6. PMID:8871560
- ↑ Naumann TA, Reznikoff WS. Tn5 transposase active site mutants. J Biol Chem. 2002 May 17;277(20):17623-9. Epub 2002 Mar 4. PMID:11877443 doi:10.1074/jbc.M200742200
- ↑ Steiniger-White M, Bhasin A, Lovell S, Rayment I, Reznikoff WS. Evidence for "unseen" transposase--DNA contacts. J Mol Biol. 2002 Oct 4;322(5):971-82. PMID:12367522
- ↑ Klenchin VA, Czyz A, Goryshin IY, Gradman R, Lovell S, Rayment I, Reznikoff WS. Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif. Nucleic Acids Res. 2008 Oct;36(18):5855-62. Epub 2008 Sep 12. PMID:18790806 doi:10.1093/nar/gkn577
|