|
|
| (69 intermediate revisions not shown.) |
| Line 1: |
Line 1: |
| - | TMPRSS2 is a membrane protein belonging to the '''type II transmembrane serine protease (TTSP) family'''. It is functionally classified as a '''trypsin-like protease''' (TLP). <ref>DOI 10.3389/fmolb.2021.666626</ref> Serine proteases are known to be involved in many physiological and pathological processes. | + | '''TMPRSS2''' ('''Transmembrane protease serine 2''') is a membrane protein belonging to the '''type II transmembrane serine protease (TTSP) family'''. It is functionally classified as a '''trypsin-like protease''' (TLP). <ref>DOI 10.3389/fmolb.2021.666626</ref> Serine proteases are known to be involved in many physiological and pathological processes, but the exact function remains unclear. It is involved in two main pathological processes, prostate cancer and viral entry. TMPRSS2 is expressed in several tissues regulated by androgens and undergoes autocatalytic cleavage for its activation. |
| | + | *'''Transmembrane protease serine 2''' has a role in proteolytic epithelial sodium channel activation<ref>PMID:35504352</ref>. |
| | + | *'''Transmembrane protease serine 11e''' is induced during inflammation response<ref>PMID:36413750</ref>. |
| | + | *'''Transmembrane protease serine 13''' facilitates virus entry into host cell by proteolytically cleaving and activating viral envelope glycoproteins. |
| | + | *'''Transmembrane protease serine 15''' converts pancreatic trypsinogen to trypsin. |
| | | | |
| - | <StructureSection load='7meq' size='340' side='right' caption='Crystal structure of human TMPRSS2 in complex with Nafamostat' scene=''> | + | <StructureSection load='7meq' size='340' side='right' caption='Glycosylated human TMPRSS2 in complex with Nafamostat (PDB code [[7meq]])' scene='89/897681/Colors/1'> |
| | | | |
| | == Protease activity == | | == Protease activity == |
| | | | |
| - | TMPRSS2, as a serine protease, cleaves peptide bonds present after positively charged residues (lysine or arginine). The main player in the catalytic mechanism is the catalytic triad formed by His296, Asp345, and Ser441. This three aminoacids are located in the active site of the enzyme. <ref>DOI 10.1073/pnas.87.17.6659</ref> | + | TMPRSS2, as a serine protease, cleaves peptide bonds present after positively charged residues (lysine or arginine). The main player in the catalytic mechanism is the catalytic triad formed by His296, Asp345, and Ser441. These three aminoacids are located in the active site of the enzyme. <ref>DOI 10.1073/pnas.87.17.6659</ref> |
| | | | |
| | The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. <ref>DOI 10.1016/j.ejps.2020.105495</ref> | | The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. <ref>DOI 10.1016/j.ejps.2020.105495</ref> |
| | + | |
| | + | == Structure == |
| | + | |
| | + | === Gene === |
| | + | |
| | + | The ''TMPRSS2'' gene resides on chromosome 21 at the band 21q22.3, extends aproximately 43.59 kb and is split into 14 exons. |
| | + | |
| | + | This gene is conserved in a wide variety of animals, such as chimpanzee, Rhesus monkey, dog, cow, mouse, rat, chicken, zebrafish, ''Caenorhabditis elegans'' and frog. It presents two alternative splicing variants resulting in a 3.25 kb and 3.21 kb transcripts, respectively. |
| | + | |
| | + | === Protein === |
| | + | |
| | + | TMPRSS2 is a 492 amino acid single-pass type II membrane protein. This protein is defined by the presence of an N-terminal cytoplasmic domain, a transmembrane helical domain (aa 84-106), and three extracellular domains: <ref>DOI 10.1136/jclinpath-2020-206987</ref> <scene name='89/897681/Domains/2'>TMPRSS2 protein domains</scene> |
| | + | *Low-density lipoprotein (LDL)-receptor class A domain: which forms a binding site for calcium. Expands from amino acid 113 to 148. |
| | + | *Scavenger receptor cysteine-rich domain (SRCR) of group A from amino acid 149 to 242. |
| | + | *Peptidase S1 domain, also known as serine protease domain (SPD) from amino acid 255 to 492. |
| | + | [[Image:TMPRSS2.jpg|thumb|left|500px|Schematic representation of TMPRSS2 structure: TM is the transmembrane domain, LDL receptor class A domain and SRCR (scavenger receptor cysteine rich domain) form the stem region. The serine protease domain is composed by the catalytic amino acid triad histidine, aspartic acid and serine, essential for proteolytic activity.]] |
| | + | |
| | + | {{Clear}} |
| | + | |
| | + | == Expression == |
| | + | |
| | + | TMPRSS2 is predominantly expressed in prostate, with relatively lower level of expression in type II pneumocytes in lungs, colon, small intestine, stomach, salivary glands, liver, kidneys and pancreas. <ref>DOI </ref> |
| | + | |
| | + | ===Autocatalytic cleavage=== |
| | + | |
| | + | As TMPRSS2 is synthesized as a single-chain proenzyme, or zymogen, it requires cleavage at a conserved Arg255-Ile256 peptide bond within its SRQSR255↓IVGGE activation motif (cleavage site denoted with an arrow) to achieve full maturation of its enzymatic activity. <ref>PMID 11245484</ref> This '''autocatalytic''' cleavage activates the 492-residue long TMPRSS2 zymogen. This modification enables the binding of Ile256 into a putative allosteric pocket ('''A-pocket'''), which induces a conformational rearrangement of the catalytic site. <ref>DOI 10.1128/jvi.00239-10</ref> |
| | + | |
| | + | After the cleavage, TMPRSS2 remains bound to the transmembrane N-terminal domains by a conserved disulfide bond, although a small fraction of the protein can be detected into the extracellular environment, the protease domain of this protein is thought to be cleaved and secreted into cell media after autocleavage.<ref>DOI 10.1016/j.biochi.2017.07.016</ref> |
| | + | |
| | + | ===Regulation=== |
| | + | |
| | + | The human TMPRSS2 gene promoter has a 15-bp androgen response element. The upregulation of TMPRSS2 mRNA by androgens appears to be mediated by the androgen receptor. |
| | | | |
| | == General function == | | == General function == |
| - | In terms of normal function, TMPRSS2 has | + | |
| - | been associated with physiological and pathological processes such as digestion, tissue remodelling, blood coagulation, fertility, inflammatory | + | In terms of normal function, TMPRSS2 has been associated with physiological and pathological processes such as digestion, tissue remodelling, blood coagulation, fertility, inflammatory responses, tumour cell invasion, apoptosis and pain. <ref>DOI 10.1097/j.pain.0000000000000130</ref> In the lung, it has been suggested that it regulates epithelial sodium currents through proteolytic cleavage of the epithelial sodium channel. However, knock out mice showed no obvious phenotypic abnormality such as death, infertility or visible sickness, and the exact physiological function of TMPRSS2 in vivo remains unknown. It is speculated that TMPRSS2 may contribute to a specialised but non vital function. <ref>DOI 10.1128/MCB.26.3.965-975.2006 </ref> |
| - | responses, tumour cell invasion, apoptosis and | + | |
| - | pain. <ref>DOI 10.1097/j.pain.0000000000000130</ref> | + | == Clinical relevance == |
| | | | |
| | === Prostate cancer === | | === Prostate cancer === |
| Line 19: |
Line 55: |
| | Prostate cancer (PC) is the most common form of cancer found in American men and the second leading cause of cancer death. <ref>DOI 10.4172/1948-5956.1000119</ref> This means that approximately 28.5% of cancers and 3.5% of cancer related deaths in men are due to PC. | | Prostate cancer (PC) is the most common form of cancer found in American men and the second leading cause of cancer death. <ref>DOI 10.4172/1948-5956.1000119</ref> This means that approximately 28.5% of cancers and 3.5% of cancer related deaths in men are due to PC. |
| | | | |
| - | The most prevalent chromosomal aberration causing this pathology is the fusion of the the promoter of transmembrane protease serine 2 (TMPRSS2) gene and the coding sequence of the erythroblastosis virus E26 (Ets) gene family members. <ref>DOI 10.1126/science.1117679</ref> Ets family members are oncogenic transcription factors. <ref>DOI 10.1038/sj.onc.1201868</ref> Therefore, the fusion of these genes leads to the production of Ets transcription factors under the control of the androgen sensitive promoter elements of TMPRSS2. Specifically, the TMPRSS2-ERG fusion has been identified in approximately 50% of PC cases. <ref>DOI 10.1016/j.ccr.2010.03.018</ref> | + | The most prevalent chromosomal aberration causing this pathology is the fusion of the the promoter of transmembrane protease serine 2 (TMPRSS2) gene and the coding sequence of the erythroblastosis virus E26 (Ets) gene family members. <ref>DOI 10.1126/science.1117679</ref> Ets family members are oncogenic transcription factors. <ref>DOI 10.1038/sj.onc.1201868</ref> Therefore, the fusion of these genes leads to the production of Ets transcription factors under the control of the androgen sensitive promoter elements of TMPRSS2. Specifically, the TMPRSS2-ERG fusion has been identified in approximately 50% of PC cases, responsible for driving carcinogenesis. <ref>DOI 10.1016/j.ccr.2010.03.018</ref> |
| | | | |
| | This mutation occurs through chromosomal translocation or intergenic deletion, with both genes on the same arm of chromosome 21, and results in overexpression of chimeric mRNA of ERG in response to androgens. There is impairment of apoptosis in TMPRSS2-ERG positive cancer cells, possibly due to disruption of the intracellular death domain or decoy receptors. <ref>DOI 10.1186/1475-2867-14-34 </ref> | | This mutation occurs through chromosomal translocation or intergenic deletion, with both genes on the same arm of chromosome 21, and results in overexpression of chimeric mRNA of ERG in response to androgens. There is impairment of apoptosis in TMPRSS2-ERG positive cancer cells, possibly due to disruption of the intracellular death domain or decoy receptors. <ref>DOI 10.1186/1475-2867-14-34 </ref> |
| Line 27: |
Line 63: |
| | === Viral entry === | | === Viral entry === |
| | | | |
| - | '''TMPRSS2''' facilitates the entry of viruses into host cells by proteolytically cleaving and activating viral envelope glycoproteins. As human TMPRSS2 is expressed in cells of the respiratory tracts, in addition to the epithelia of the gastrointestinal and urogenital systems, it mediates the entry of several viruses related to respiratory diseases into the host cells, including Influenza virus and the human coronaviruses HCoV-229E, MERS-CoV, SARS-CoV and SARS-CoV-2 (COVID-19 virus).
| + | TMPRSS2 facilitates the entry of viruses into host cells by proteolytically cleaving and activating viral envelope glycoproteins (viral spike protein). As human TMPRSS2 is expressed in cells of the respiratory tracts, in addition to the epithelia of the gastrointestinal and urogenital systems, it mediates the entry of several viruses related to respiratory diseases into the host cells, including Influenza virus and the human coronaviruses HCoV-229E, MERS-CoV, SARS-CoV and SARS-CoV-2 (COVID-19 virus) <ref>DOI 10.1136/jclinpath-2020-206987</ref>. |
| | + | [[Image:viral entry.jpg|thumb|left|400px|Viral entry mediated by TMPRSS2.]] |
| | + | |
| | + | {{Clear}} |
| | | | |
| | ====SARS-CoV-2==== | | ====SARS-CoV-2==== |
| - | SARS-CoV-2 entry is achieved by a receptor-mediated endocytosis pathway in which the spike (S) glycoprotein, located on the outer envelope of the virus, interacts with the host angiotensin-converting enzyme 2 (ACE2), a receptor located in the surface of host cells, which allows the virus to infect cells. Prior to this interaction, S protein is needed to be cleaved by different protease enzymes (furins, cathepsins, serine proteases). In this respect, cleavage of S protein by '''TMPRSS2''' is preferred for ''Coronaviridae'' family infection over other proteases, such as the endosomal cathepsins. | |
| | | | |
| - | == Structure ==
| + | SARS-CoV-2 entry is achieved by a receptor-mediated endocytosis pathway in which the spike (S) glycoprotein, located on the outer envelope of the virus, interacts with the host angiotensin-converting enzyme 2 (ACE2), a receptor located in the surface of host cells, which allows the virus to infect cells. Prior to this interaction, S protein is needed to be cleaved by different protease enzymes (furins, cathepsins, serine proteases) <ref>DOI 10.1128/AAC.00754-20</ref>. SARS-CoV-2 S protein presents two functional domains S1, the receptor binding domain, and S2, that contains functional elements involved in membrane fusion. There are multiple sites in which this protein can be cleaved; one of these is at the S1/S2 boundary and another within S2. The S1/S2 cleavage site contains multiple arginine residues, which allows the action of serine proteases <ref>DOI 10.1136/jclinpath-2020-206987</ref>. ''Coronaviridae'' family tend to prefer "TMPRSS2" for the cleavage of S protein over other proteases, such as the endosomal cathepsins. As this protease is expressed in SARS-CoV-2 target cells throughout the human respiratory tract, it is also required for the spread of this virus. However, it has been demonstrated to be dispensable for the development or homeostasis of mice models, so it can be considered a potential target to fight the infection of these viruses. |
| | | | |
| - | === Gene ===
| + | [[Image:CLEAVAGE.jpg|thumb|left|300px|Schematic representation of the cleavage sites in TMPRSS2 protein structure.<ref>DOI 10.1016/j.biochi.2017.07.016</ref>]] |
| | | | |
| - | The ''TMPRSS2'' gene resides on chromosome 21 at the band 21q22.3, extends aproximately 43.59 kb and is split into 14 exons.
| + | {{Clear}} |
| | | | |
| - | This gene is conserved in a wide variety of animals, such as chimpanzee, Rhesus monkey, dog, cow, mouse, rat, chicken, zebrafish, ''Caenorhabditis elegans'' and frog.
| + | == Pharmacological therapeutic approaches == |
| | | | |
| - | This gene presents two alternative splicing variants resulting in a 3.25 kb and 3.21 kb transcripts, respectively.
| + | === Inhibitors of TMPRSS2 === |
| | | | |
| - | === Protein === | + | ==== Nafamostat mesylate ==== |
| | | | |
| - | TMPRSS2 is a 492 amino acid single-pass type II membrane protein. This protein is defined by the presence of an N-terminal cytoplasmic domain, a transmembrane helical domain, and three extracellular domains: <ref>DOI 10.1136/jclinpath-2020-206987</ref>
| + | Nafamostat mesylate (FUT-175; CAS number: 81525-10-2) is an artificial serine protease inhibitor clinically approved in Japan for the treatment of acute pancreatitis, intravascular coagulation dissemination, and extracorporeal circulation antioxidation. <ref>DOI 10.3390/cells9071652</ref> |
| - | *Low-density lipoprotein (LDL)-receptor class A domain: which forms a binding site for calcium
| + | |
| - | *Scavenger receptor cysteine-rich domain (SRCR)
| + | |
| - | *Peptidase S1 domain, also known as serine protease domain (SPD)
| + | |
| | | | |
| - | [[Image:TMPRSS2.jpg]]
| + | Although nafamostat potently neutralizes TMPRSS2 activity, it is non-selective and disables trypsin-like serine proteases involved in coagulation such as plasmin, FXa, and FXIIa, as well as other TTSPs through its generic arginine-like engagement with the S1 subsite.<ref>DOI 10.1101/2020.06.23.167544</ref><ref>DOI 10.1159/000215139</ref> Also, it requires continuous intravenous infusion to approach therapeutic concentrations for COVID-19 owing to its short biological half-life of 8 minutes. |
| | | | |
| - | == Expression ==
| + | [[Image:ACYL.jpg|thumb|left|500px|Close-up view of the SP catalytic triad residues and the post-activation Asp440:Ile256 salt bridge showing complete maturation of the protease. Nafamostat treatment results in phenylguanidino acylation of Ser441. Polar contacts are shown as yellow dashed lines. <ref>DOI 10.1101/2021.06.23.449282</ref>]] |
| | | | |
| - | == Pharmacological therapeutic approaches ==
| + | {{Clear}} |
| | | | |
| - | === Inhibitors of TMPRSS2 === | + | ==== Camostat mesylate ==== |
| - | ==== Nafamostat mesylate ====
| + | |
| | | | |
| - | Nafamostat mesylate (FUT-175; CAS number: 81525-10-2) is an artificial serine protease inhibitor clinically approved in Japan for the treatment of acute pancreatitis, intravascular coagulation dissemination, and extracorporeal circulation antioxidation. <ref>DOI 10.3390/cells9071652</ref>
| + | Camostat is a serine protease inhibitor used in the treatment of cancer, pancreatitis and liver fibrosis. A recent study demonstrated that the inhibition of TMPRSS2 with camostat led to a 10-fold reduction in SARS-CoV titers in Calu-3 cells. |
| | | | |
| - | This drug is a competitive inhibitor of the binding active site, as well as Camostat. Both are reactive esters that form the same slowly-reversible phenylguanidino covalent complex with the catalytic serine (Ser441) residue of trypsin-like serine proteases.<ref>DOI 10.1101/2021.06.23.449282</ref>
| + | ==== Nafamostat vs. Camostat mesylate ==== |
| | | | |
| - | Nafamostat demonstrated enhanced potency over camostat with IC50 values of (1.7±0.2) and (17±4) nM, respectively. | + | Nafamostat and Camostat are competitive inhibitors of the binding active site. They are both reactive esters that form the same slowly-reversible phenylguanidino covalent complex with the catalytic serine (Ser441) residue of trypsin-like serine proteases.<ref>DOI 10.1101/2021.06.23.449282</ref> However, Nafamostat demonstrated enhanced potency over camostat with IC50 values of (1.7±0.2) and (17±4) nM, respectively. Nafamostat has demonstrated being able to block MERS-CoV infection in vitro via inhabiting the activity of TMPRSS2, and reduce viral entry by 100-fold at a concentration of as low as 1 nM, which is more effective than camostat. |
| | | | |
| - | Although nafamostat potently neutralizes TMPRSS2 activity, it is non-selective and disables trypsin-like serine proteases involved in coagulation such as plasmin, FXa, and FXIIa, as well as other TTSPs through its generic arginine-like engagement with the S1 subsite.<ref>DOI 10.1101/2020.06.23.167544</ref><ref>DOI 10.1159/000215139</ref>
| + | [[Image:Viability.jpg|thumb|left|500px|Cell viability of Calu3 cell line treated with Gabexate (grey), Nafamostat (red) and Camostat (pink). <ref>DOI 10.1128/AAC.00754-20</ref>]][[Image:Nafamostat vs Camostat.jpg|thumb|left|500px|Entry efficiency of SARS-2-S in Calu3 cell line treated with Gabexate (grey), Nafamostat (red) and Camostat (pink). <ref>DOI 10.1128/AAC.00754-20</ref>]] |
| | | | |
| - | Also it requires continuous intravenous infusion to approach therapeutic concentrations for COVID-19 owing to its short biological half-life of 8 minutes.
| + | {{Clear}} |
| | | | |
| - | [[Image:ACYL.jpg]] | |
| - | | |
| - | ==== Camostat mesylate ==== | |
| | ==== Bromhexine ==== | | ==== Bromhexine ==== |
| | | | |
| - | Repurposing of the mucolytic agent called bromhexine, a TMPRSS2 inhibitor, has been also proposed for COVID-19 therapy.<ref>DOI 10.1016/j.phrs.2020.104837</ref>
| + | Bromhexine is a mucolytic agent that acts as a TMPRSS2 inhibitor. Given that BHH is an FDA approved drug with no significant adverse effects, it could be used for treatment of influenza virus and coronavirus infections. However, recent studies <ref>DOI 10.1101/2020.06.23.167544</ref> <ref>DOI 10.1101/2021.06.23.449282</ref> have shown this drug has no effectiveness in blocking SARS-CoV-2 pseudovirus entry and further research is needed regarding the design of novel and more selective TMPRSS2 inhibitors. It has also been proposed for attenuation of prostate cancer in mice.<ref>DOI 10.1016/j.phrs.2020.104837</ref> |
| - | | + | [[Image:Bromhexine.jpg|thumb|left|500px|TMPRSS2 peptidase activity blocked by clinical protease inhibitors, with no inhibition seen for bromhexine.<ref>DOI 10.1101/2021.06.23.449282</ref>]] |
| - | ==== Peptidomimetics ====
| + | |
| | | | |
| | + | {{Clear}} |
| | | | |
| | === Transcriptional inhibition === | | === Transcriptional inhibition === |
| Line 83: |
Line 114: |
| | | | |
| | As TMPRSS2 expression seems to be modulated by estrogens and androgens, data suggest that the activation of estrogen pathways or inhibition of androgen pathways may be a new target for therapeutic clinical intervention for symptom amelioration in COVID-19 patients. <ref>DOI 10.20944/preprints202003.0360.v2 | | As TMPRSS2 expression seems to be modulated by estrogens and androgens, data suggest that the activation of estrogen pathways or inhibition of androgen pathways may be a new target for therapeutic clinical intervention for symptom amelioration in COVID-19 patients. <ref>DOI 10.20944/preprints202003.0360.v2 |
| - | </ref> | + | </ref> In fact, according to this same study estrogen-related compounds and androgen receptor antagonists appear to be the most securely identified down-regulators of TMPRSS2 expression amongst FDA approved drugs and other widely tested compounds. |
| | | | |
| - | Using computational and experimental methods, estrogen and androgen-related compounds such as genistein, estradiol, and enzatulamide have been shown to reduce TMPRSS2 expression.
| + | [[Image:COMPUESTOS.jpg|thumb|left|500px|TMPRSS2 modulators: (A) chemical structure of TMPRSS2 inhibitors and (B) some transcriptional inhibitors. <ref>DOI 10.1021/acs.jmedchem.0c00606</ref>]] |
| | | | |
| - | [[Image:COMPUESTOS.jpg]]
| + | {{Clear}} |
| | | | |
| | + | ==3D structures of transmembrane protease serine== |
| | | | |
| | + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} |
| | | | |
| - | == Structural highlights ==
| + | [[7meq]], [[7xyd]], [[7y0e]] – hTMPRRS2 + drug – human<br /> |
| | + | [[7y0f]], [[8hd8]] – hTMPRRS2 + inhibitor<br /> |
| | + | [[2oq5]] – hTMPRRS11e + peptide inhibitor <br /> |
| | + | [[6kd5]] – hTMPRRS13 + peptide inhibitor <br /> |
| | + | [[7wqx]] – hTMPRRS15 – Cryo EM <br /> |
| | | | |
| - | | |
| - | | |
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | |
| - | | |
| - | </StructureSection> | |
| | == References == | | == References == |
| | <references/> | | <references/> |
| | + | </StructureSection> |
| | + | [[Category:Topic Page]] |
|
Protease activity
TMPRSS2, as a serine protease, cleaves peptide bonds present after positively charged residues (lysine or arginine). The main player in the catalytic mechanism is the catalytic triad formed by His296, Asp345, and Ser441. These three aminoacids are located in the active site of the enzyme. [4]
The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. [5]
Structure
Gene
The TMPRSS2 gene resides on chromosome 21 at the band 21q22.3, extends aproximately 43.59 kb and is split into 14 exons.
This gene is conserved in a wide variety of animals, such as chimpanzee, Rhesus monkey, dog, cow, mouse, rat, chicken, zebrafish, Caenorhabditis elegans and frog. It presents two alternative splicing variants resulting in a 3.25 kb and 3.21 kb transcripts, respectively.
Protein
TMPRSS2 is a 492 amino acid single-pass type II membrane protein. This protein is defined by the presence of an N-terminal cytoplasmic domain, a transmembrane helical domain (aa 84-106), and three extracellular domains: [6]
- Low-density lipoprotein (LDL)-receptor class A domain: which forms a binding site for calcium. Expands from amino acid 113 to 148.
- Scavenger receptor cysteine-rich domain (SRCR) of group A from amino acid 149 to 242.
- Peptidase S1 domain, also known as serine protease domain (SPD) from amino acid 255 to 492.
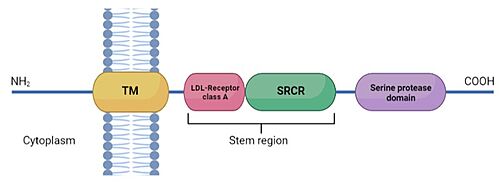 Schematic representation of TMPRSS2 structure: TM is the transmembrane domain, LDL receptor class A domain and SRCR (scavenger receptor cysteine rich domain) form the stem region. The serine protease domain is composed by the catalytic amino acid triad histidine, aspartic acid and serine, essential for proteolytic activity. Expression
TMPRSS2 is predominantly expressed in prostate, with relatively lower level of expression in type II pneumocytes in lungs, colon, small intestine, stomach, salivary glands, liver, kidneys and pancreas. [7]
Autocatalytic cleavage
As TMPRSS2 is synthesized as a single-chain proenzyme, or zymogen, it requires cleavage at a conserved Arg255-Ile256 peptide bond within its SRQSR255↓IVGGE activation motif (cleavage site denoted with an arrow) to achieve full maturation of its enzymatic activity. [8] This autocatalytic cleavage activates the 492-residue long TMPRSS2 zymogen. This modification enables the binding of Ile256 into a putative allosteric pocket (A-pocket), which induces a conformational rearrangement of the catalytic site. [9]
After the cleavage, TMPRSS2 remains bound to the transmembrane N-terminal domains by a conserved disulfide bond, although a small fraction of the protein can be detected into the extracellular environment, the protease domain of this protein is thought to be cleaved and secreted into cell media after autocleavage.[10]
Regulation
The human TMPRSS2 gene promoter has a 15-bp androgen response element. The upregulation of TMPRSS2 mRNA by androgens appears to be mediated by the androgen receptor.
General function
In terms of normal function, TMPRSS2 has been associated with physiological and pathological processes such as digestion, tissue remodelling, blood coagulation, fertility, inflammatory responses, tumour cell invasion, apoptosis and pain. [11] In the lung, it has been suggested that it regulates epithelial sodium currents through proteolytic cleavage of the epithelial sodium channel. However, knock out mice showed no obvious phenotypic abnormality such as death, infertility or visible sickness, and the exact physiological function of TMPRSS2 in vivo remains unknown. It is speculated that TMPRSS2 may contribute to a specialised but non vital function. [12]
Clinical relevance
Prostate cancer
Prostate cancer (PC) is the most common form of cancer found in American men and the second leading cause of cancer death. [13] This means that approximately 28.5% of cancers and 3.5% of cancer related deaths in men are due to PC.
The most prevalent chromosomal aberration causing this pathology is the fusion of the the promoter of transmembrane protease serine 2 (TMPRSS2) gene and the coding sequence of the erythroblastosis virus E26 (Ets) gene family members. [14] Ets family members are oncogenic transcription factors. [15] Therefore, the fusion of these genes leads to the production of Ets transcription factors under the control of the androgen sensitive promoter elements of TMPRSS2. Specifically, the TMPRSS2-ERG fusion has been identified in approximately 50% of PC cases, responsible for driving carcinogenesis. [16]
This mutation occurs through chromosomal translocation or intergenic deletion, with both genes on the same arm of chromosome 21, and results in overexpression of chimeric mRNA of ERG in response to androgens. There is impairment of apoptosis in TMPRSS2-ERG positive cancer cells, possibly due to disruption of the intracellular death domain or decoy receptors. [17]
The high prevalence of these gene fusions, in particular TMPRSS2-ERG, makes them attractive as potential diagnostic and prognostic indicators, as well as making them a potential target for tailored therapies.
Viral entry
TMPRSS2 facilitates the entry of viruses into host cells by proteolytically cleaving and activating viral envelope glycoproteins (viral spike protein). As human TMPRSS2 is expressed in cells of the respiratory tracts, in addition to the epithelia of the gastrointestinal and urogenital systems, it mediates the entry of several viruses related to respiratory diseases into the host cells, including Influenza virus and the human coronaviruses HCoV-229E, MERS-CoV, SARS-CoV and SARS-CoV-2 (COVID-19 virus) [18].
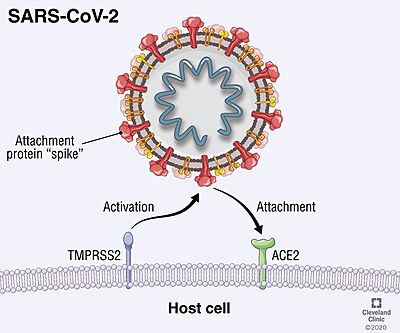 Viral entry mediated by TMPRSS2. SARS-CoV-2
SARS-CoV-2 entry is achieved by a receptor-mediated endocytosis pathway in which the spike (S) glycoprotein, located on the outer envelope of the virus, interacts with the host angiotensin-converting enzyme 2 (ACE2), a receptor located in the surface of host cells, which allows the virus to infect cells. Prior to this interaction, S protein is needed to be cleaved by different protease enzymes (furins, cathepsins, serine proteases) [19]. SARS-CoV-2 S protein presents two functional domains S1, the receptor binding domain, and S2, that contains functional elements involved in membrane fusion. There are multiple sites in which this protein can be cleaved; one of these is at the S1/S2 boundary and another within S2. The S1/S2 cleavage site contains multiple arginine residues, which allows the action of serine proteases [20]. Coronaviridae family tend to prefer "TMPRSS2" for the cleavage of S protein over other proteases, such as the endosomal cathepsins. As this protease is expressed in SARS-CoV-2 target cells throughout the human respiratory tract, it is also required for the spread of this virus. However, it has been demonstrated to be dispensable for the development or homeostasis of mice models, so it can be considered a potential target to fight the infection of these viruses.
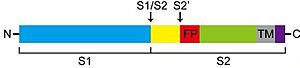 Schematic representation of the cleavage sites in TMPRSS2 protein structure. [21] Pharmacological therapeutic approaches
Inhibitors of TMPRSS2
Nafamostat mesylate
Nafamostat mesylate (FUT-175; CAS number: 81525-10-2) is an artificial serine protease inhibitor clinically approved in Japan for the treatment of acute pancreatitis, intravascular coagulation dissemination, and extracorporeal circulation antioxidation. [22]
Although nafamostat potently neutralizes TMPRSS2 activity, it is non-selective and disables trypsin-like serine proteases involved in coagulation such as plasmin, FXa, and FXIIa, as well as other TTSPs through its generic arginine-like engagement with the S1 subsite.[23][24] Also, it requires continuous intravenous infusion to approach therapeutic concentrations for COVID-19 owing to its short biological half-life of 8 minutes.
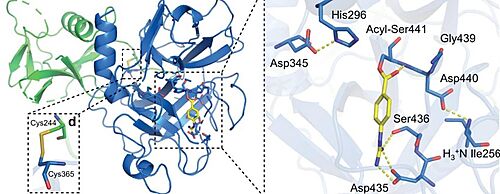 Close-up view of the SP catalytic triad residues and the post-activation Asp440:Ile256 salt bridge showing complete maturation of the protease. Nafamostat treatment results in phenylguanidino acylation of Ser441. Polar contacts are shown as yellow dashed lines. [25] Camostat mesylate
Camostat is a serine protease inhibitor used in the treatment of cancer, pancreatitis and liver fibrosis. A recent study demonstrated that the inhibition of TMPRSS2 with camostat led to a 10-fold reduction in SARS-CoV titers in Calu-3 cells.
Nafamostat vs. Camostat mesylate
Nafamostat and Camostat are competitive inhibitors of the binding active site. They are both reactive esters that form the same slowly-reversible phenylguanidino covalent complex with the catalytic serine (Ser441) residue of trypsin-like serine proteases.[26] However, Nafamostat demonstrated enhanced potency over camostat with IC50 values of (1.7±0.2) and (17±4) nM, respectively. Nafamostat has demonstrated being able to block MERS-CoV infection in vitro via inhabiting the activity of TMPRSS2, and reduce viral entry by 100-fold at a concentration of as low as 1 nM, which is more effective than camostat.
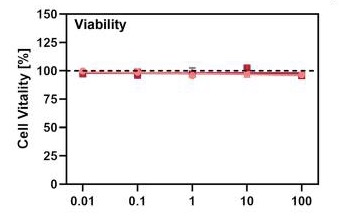 Cell viability of Calu3 cell line treated with Gabexate (grey), Nafamostat (red) and Camostat (pink). [27]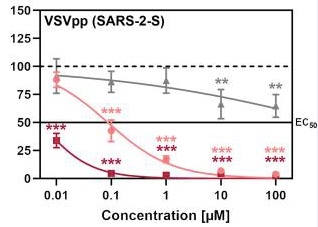 Entry efficiency of SARS-2-S in Calu3 cell line treated with Gabexate (grey), Nafamostat (red) and Camostat (pink). [28] Bromhexine
Bromhexine is a mucolytic agent that acts as a TMPRSS2 inhibitor. Given that BHH is an FDA approved drug with no significant adverse effects, it could be used for treatment of influenza virus and coronavirus infections. However, recent studies [29] [30] have shown this drug has no effectiveness in blocking SARS-CoV-2 pseudovirus entry and further research is needed regarding the design of novel and more selective TMPRSS2 inhibitors. It has also been proposed for attenuation of prostate cancer in mice.[31]
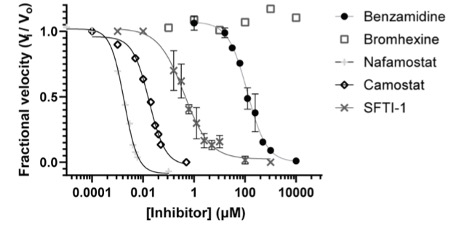 TMPRSS2 peptidase activity blocked by clinical protease inhibitors, with no inhibition seen for bromhexine. [32] Transcriptional inhibition
Transcriptional inhibition of TMPRSS2 has been proposed as a new therapeutic option.
As TMPRSS2 expression seems to be modulated by estrogens and androgens, data suggest that the activation of estrogen pathways or inhibition of androgen pathways may be a new target for therapeutic clinical intervention for symptom amelioration in COVID-19 patients. [33] In fact, according to this same study estrogen-related compounds and androgen receptor antagonists appear to be the most securely identified down-regulators of TMPRSS2 expression amongst FDA approved drugs and other widely tested compounds.
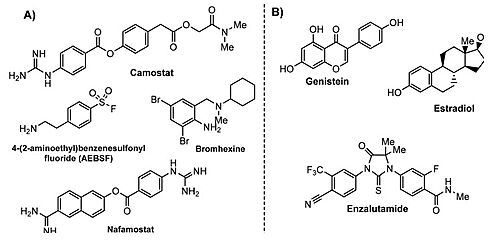 TMPRSS2 modulators: (A) chemical structure of TMPRSS2 inhibitors and (B) some transcriptional inhibitors. [34] 3D structures of transmembrane protease serine
Updated on 20-August-2024
7meq, 7xyd, 7y0e – hTMPRRS2 + drug – human
7y0f, 8hd8 – hTMPRRS2 + inhibitor
2oq5 – hTMPRRS11e + peptide inhibitor
6kd5 – hTMPRRS13 + peptide inhibitor
7wqx – hTMPRRS15 – Cryo EM
References
- ↑ Sgrignani J, Cavalli A. Computational Identification of a Putative Allosteric Binding Pocket in TMPRSS2. Front Mol Biosci. 2021 Apr 30;8:666626. doi: 10.3389/fmolb.2021.666626., eCollection 2021. PMID:33996911 doi:http://dx.doi.org/10.3389/fmolb.2021.666626
- ↑ Sure F, Bertog M, Afonso S, Diakov A, Rinke R, Madej MG, Wittmann S, Gramberg T, Korbmacher C, Ilyaskin AV. Transmembrane serine protease 2 (TMPRSS2) proteolytically activates the epithelial sodium channel (ENaC) by cleaving the channel's γ-subunit. J Biol Chem. 2022 Jun;298(6):102004. PMID:35504352 doi:10.1016/j.jbc.2022.102004
- ↑ Zhang W, Chen Z, Wang T, Wang X, Liu L, Huang W, Li S. Increased Expression of TMPRSS11E Is Involved in LPS Inflammation. Am J Respir Cell Mol Biol. 2023 Apr;68(4):406-416. PMID:36413750 doi:10.1165/rcmb.2022-0256OC
- ↑ Evnin LB, Vasquez JR, Craik CS. Substrate specificity of trypsin investigated by using a genetic selection. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6659-63. doi: 10.1073/pnas.87.17.6659. PMID:2204062 doi:http://dx.doi.org/10.1073/pnas.87.17.6659
- ↑ Singh N, Decroly E, Khatib AM, Villoutreix BO. Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages. Eur J Pharm Sci. 2020 Oct 1;153:105495. doi: 10.1016/j.ejps.2020.105495. Epub, 2020 Jul 28. PMID:32730844 doi:http://dx.doi.org/10.1016/j.ejps.2020.105495
- ↑ Thunders M, Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J Clin Pathol. 2020 Dec;73(12):773-776. doi: 10.1136/jclinpath-2020-206987. Epub , 2020 Sep 1. PMID:32873700 doi:http://dx.doi.org/10.1136/jclinpath-2020-206987
- ↑ . PMID:216315890657
- ↑ Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB, Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001 Feb 15;61(4):1686-92. PMID:11245484
- ↑ Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, Steffen I, Choi SY, Park Y, Schneider H, Schughart K, Pohlmann S. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol. 2010 Oct;84(19):10016-25. doi: 10.1128/JVI.00239-10. Epub 2010 Jul 14. PMID:20631123 doi:http://dx.doi.org/10.1128/JVI.00239-10
- ↑ Shen LW, Mao HJ, Wu YL, Tanaka Y, Zhang W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017 Nov;142:1-10. doi: 10.1016/j.biochi.2017.07.016. Epub 2017 Aug 1. PMID:28778717 doi:http://dx.doi.org/10.1016/j.biochi.2017.07.016
- ↑ Lam DK, Dang D, Flynn AN, Hardt M, Schmidt BL. TMPRSS2, a novel membrane-anchored mediator in cancer pain. Pain. 2015 May;156(5):923-930. doi: 10.1097/j.pain.0000000000000130. PMID:25734995 doi:http://dx.doi.org/10.1097/j.pain.0000000000000130
- ↑ Kim TS, Heinlein C, Hackman RC, Nelson PS. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol Cell Biol. 2006 Feb;26(3):965-75. doi: 10.1128/MCB.26.3.965-975.2006. PMID:16428450 doi:http://dx.doi.org/10.1128/MCB.26.3.965-975.2006
- ↑ St John J, Powell K, Conley-Lacomb MK, Chinni SR. TMPRSS2-ERG Fusion Gene Expression in Prostate Tumor Cells and Its Clinical and Biological Significance in Prostate Cancer Progression. J Cancer Sci Ther. 2012 Apr 26;4(4):94-101. doi: 10.4172/1948-5956.1000119. PMID:23264855 doi:http://dx.doi.org/10.4172/1948-5956.1000119
- ↑ Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005 Oct 28;310(5748):644-8. doi: 10.1126/science.1117679. PMID:16254181 doi:http://dx.doi.org/10.1126/science.1117679
- ↑ Carrere S, Verger A, Flourens A, Stehelin D, Duterque-Coquillaud M. Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains. Oncogene. 1998 Jun 25;16(25):3261-8. doi: 10.1038/sj.onc.1201868. PMID:9681824 doi:http://dx.doi.org/10.1038/sj.onc.1201868
- ↑ Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, Cheng H, Laxman B, Vellaichamy A, Shankar S, Li Y, Dhanasekaran SM, Morey R, Barrette T, Lonigro RJ, Tomlins SA, Varambally S, Qin ZS, Chinnaiyan AM. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010 May 18;17(5):443-54. doi: 10.1016/j.ccr.2010.03.018. PMID:20478527 doi:http://dx.doi.org/10.1016/j.ccr.2010.03.018
- ↑ Farooqi AA, Hou MF, Chen CC, Wang CL, Chang HW. Androgen receptor and gene network: Micromechanics reassemble the signaling machinery of TMPRSS2-ERG positive prostate cancer cells. Cancer Cell Int. 2014 Apr 17;14:34. doi: 10.1186/1475-2867-14-34. eCollection, 2014. PMID:24739220 doi:http://dx.doi.org/10.1186/1475-2867-14-34
- ↑ Thunders M, Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J Clin Pathol. 2020 Dec;73(12):773-776. doi: 10.1136/jclinpath-2020-206987. Epub , 2020 Sep 1. PMID:32873700 doi:http://dx.doi.org/10.1136/jclinpath-2020-206987
- ↑ Hoffmann M, Schroeder S, Kleine-Weber H, Muller MA, Drosten C, Pohlmann S. Nafamostat Mesylate Blocks Activation of SARS-CoV-2: New Treatment Option for COVID-19. Antimicrob Agents Chemother. 2020 May 21;64(6). pii: AAC.00754-20. doi:, 10.1128/AAC.00754-20. Print 2020 May 21. PMID:32312781 doi:http://dx.doi.org/10.1128/AAC.00754-20
- ↑ Thunders M, Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J Clin Pathol. 2020 Dec;73(12):773-776. doi: 10.1136/jclinpath-2020-206987. Epub , 2020 Sep 1. PMID:32873700 doi:http://dx.doi.org/10.1136/jclinpath-2020-206987
- ↑ Shen LW, Mao HJ, Wu YL, Tanaka Y, Zhang W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017 Nov;142:1-10. doi: 10.1016/j.biochi.2017.07.016. Epub 2017 Aug 1. PMID:28778717 doi:http://dx.doi.org/10.1016/j.biochi.2017.07.016
- ↑ Amraei R, Rahimi N. COVID-19, Renin-Angiotensin System and Endothelial Dysfunction. Cells. 2020 Jul 9;9(7). pii: cells9071652. doi: 10.3390/cells9071652. PMID:32660065 doi:http://dx.doi.org/10.3390/cells9071652
- ↑ Shrimp JH, Kales SC, Sanderson PE, Simeonov A, Shen M, Hall MD. An Enzymatic TMPRSS2 Assay for Assessment of Clinical Candidates and Discovery of Inhibitors as Potential Treatment of COVID-19. bioRxiv. 2020 Aug 6. doi: 10.1101/2020.06.23.167544. PMID:32596694 doi:http://dx.doi.org/10.1101/2020.06.23.167544
- ↑ Hitomi Y, Ikari N, Fujii S. Inhibitory effect of a new synthetic protease inhibitor (FUT-175) on the coagulation system. Haemostasis. 1985;15(3):164-8. doi: 10.1159/000215139. PMID:3161808 doi:http://dx.doi.org/10.1159/000215139
- ↑ doi: https://dx.doi.org/10.1101/2021.06.23.449282
- ↑ doi: https://dx.doi.org/10.1101/2021.06.23.449282
- ↑ Hoffmann M, Schroeder S, Kleine-Weber H, Muller MA, Drosten C, Pohlmann S. Nafamostat Mesylate Blocks Activation of SARS-CoV-2: New Treatment Option for COVID-19. Antimicrob Agents Chemother. 2020 May 21;64(6). pii: AAC.00754-20. doi:, 10.1128/AAC.00754-20. Print 2020 May 21. PMID:32312781 doi:http://dx.doi.org/10.1128/AAC.00754-20
- ↑ Hoffmann M, Schroeder S, Kleine-Weber H, Muller MA, Drosten C, Pohlmann S. Nafamostat Mesylate Blocks Activation of SARS-CoV-2: New Treatment Option for COVID-19. Antimicrob Agents Chemother. 2020 May 21;64(6). pii: AAC.00754-20. doi:, 10.1128/AAC.00754-20. Print 2020 May 21. PMID:32312781 doi:http://dx.doi.org/10.1128/AAC.00754-20
- ↑ Shrimp JH, Kales SC, Sanderson PE, Simeonov A, Shen M, Hall MD. An Enzymatic TMPRSS2 Assay for Assessment of Clinical Candidates and Discovery of Inhibitors as Potential Treatment of COVID-19. bioRxiv. 2020 Aug 6. doi: 10.1101/2020.06.23.167544. PMID:32596694 doi:http://dx.doi.org/10.1101/2020.06.23.167544
- ↑ doi: https://dx.doi.org/10.1101/2021.06.23.449282
- ↑ Maggio R, Corsini GU. Repurposing the mucolytic cough suppressant and TMPRSS2 protease inhibitor bromhexine for the prevention and management of SARS-CoV-2 infection. Pharmacol Res. 2020 Jul;157:104837. doi: 10.1016/j.phrs.2020.104837. Epub 2020, Apr 22. PMID:32334052 doi:http://dx.doi.org/10.1016/j.phrs.2020.104837
- ↑ doi: https://dx.doi.org/10.1101/2021.06.23.449282
- ↑ doi: https://dx.doi.org/10.20944/preprints202003.0360.v2
- ↑ Gil C, Ginex T, Maestro I, Nozal V, Barrado-Gil L, Cuesta-Geijo MA, Urquiza J, Ramirez D, Alonso C, Campillo NE, Martinez A. COVID-19: Drug Targets and Potential Treatments. J Med Chem. 2020 Nov 12;63(21):12359-12386. doi: 10.1021/acs.jmedchem.0c00606., Epub 2020 Jun 26. PMID:32511912 doi:http://dx.doi.org/10.1021/acs.jmedchem.0c00606
|








