Transmembrane protease serine 2
From Proteopedia
(Difference between revisions)
| (18 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
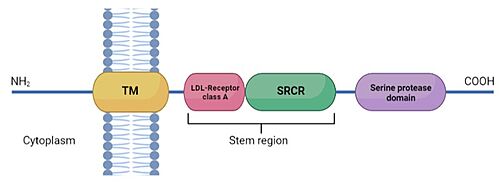
| - | TMPRSS2 is a membrane protein belonging to the '''type II transmembrane serine protease (TTSP) family'''. It is functionally classified as a '''trypsin-like protease''' (TLP). <ref>DOI 10.3389/fmolb.2021.666626</ref> Serine proteases are known to be involved in many physiological and pathological processes, but the exact function remains | + | '''TMPRSS2''' ('''Transmembrane protease serine 2''') is a membrane protein belonging to the '''type II transmembrane serine protease (TTSP) family'''. It is functionally classified as a '''trypsin-like protease''' (TLP). <ref>DOI 10.3389/fmolb.2021.666626</ref> Serine proteases are known to be involved in many physiological and pathological processes, but the exact function remains unclear. It is involved in two main pathological processes, prostate cancer and viral entry. TMPRSS2 is expressed in several tissues regulated by androgens and undergoes autocatalytic cleavage for its activation. |
| + | *'''Transmembrane protease serine 2''' has a role in proteolytic epithelial sodium channel activation<ref>PMID:35504352</ref>. | ||
| + | *'''Transmembrane protease serine 11e''' is induced during inflammation response<ref>PMID:36413750</ref>. | ||
| + | *'''Transmembrane protease serine 13''' facilitates virus entry into host cell by proteolytically cleaving and activating viral envelope glycoproteins. | ||
| + | *'''Transmembrane protease serine 15''' converts pancreatic trypsinogen to trypsin. | ||
| - | <StructureSection load='7meq' size='340' side='right' caption=' | + | <StructureSection load='7meq' size='340' side='right' caption='Glycosylated human TMPRSS2 in complex with Nafamostat (PDB code [[7meq]])' scene='89/897681/Colors/1'> |
== Protease activity == | == Protease activity == | ||
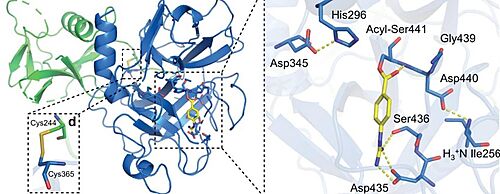
| - | TMPRSS2, as a serine protease, cleaves peptide bonds present after positively charged residues (lysine or arginine). The main player in the catalytic mechanism is the catalytic triad formed by His296, Asp345, and Ser441. | + | TMPRSS2, as a serine protease, cleaves peptide bonds present after positively charged residues (lysine or arginine). The main player in the catalytic mechanism is the catalytic triad formed by His296, Asp345, and Ser441. These three aminoacids are located in the active site of the enzyme. <ref>DOI 10.1073/pnas.87.17.6659</ref> |
The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. <ref>DOI 10.1016/j.ejps.2020.105495</ref> | The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. <ref>DOI 10.1016/j.ejps.2020.105495</ref> | ||
| Line 29: | Line 33: | ||
== Expression == | == Expression == | ||
| - | TMPRSS2 is predominantly expressed in prostate, with relatively lower level of expression in type II pneumocytes in lungs, colon, small intestine, stomach, salivary glands, liver, kidneys and | + | TMPRSS2 is predominantly expressed in prostate, with relatively lower level of expression in type II pneumocytes in lungs, colon, small intestine, stomach, salivary glands, liver, kidneys and pancreas. <ref>DOI </ref> |
===Autocatalytic cleavage=== | ===Autocatalytic cleavage=== | ||
| Line 68: | Line 72: | ||
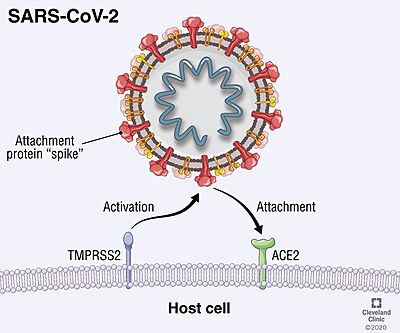
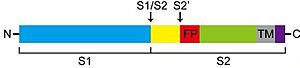
SARS-CoV-2 entry is achieved by a receptor-mediated endocytosis pathway in which the spike (S) glycoprotein, located on the outer envelope of the virus, interacts with the host angiotensin-converting enzyme 2 (ACE2), a receptor located in the surface of host cells, which allows the virus to infect cells. Prior to this interaction, S protein is needed to be cleaved by different protease enzymes (furins, cathepsins, serine proteases) <ref>DOI 10.1128/AAC.00754-20</ref>. SARS-CoV-2 S protein presents two functional domains S1, the receptor binding domain, and S2, that contains functional elements involved in membrane fusion. There are multiple sites in which this protein can be cleaved; one of these is at the S1/S2 boundary and another within S2. The S1/S2 cleavage site contains multiple arginine residues, which allows the action of serine proteases <ref>DOI 10.1136/jclinpath-2020-206987</ref>. ''Coronaviridae'' family tend to prefer "TMPRSS2" for the cleavage of S protein over other proteases, such as the endosomal cathepsins. As this protease is expressed in SARS-CoV-2 target cells throughout the human respiratory tract, it is also required for the spread of this virus. However, it has been demonstrated to be dispensable for the development or homeostasis of mice models, so it can be considered a potential target to fight the infection of these viruses. | SARS-CoV-2 entry is achieved by a receptor-mediated endocytosis pathway in which the spike (S) glycoprotein, located on the outer envelope of the virus, interacts with the host angiotensin-converting enzyme 2 (ACE2), a receptor located in the surface of host cells, which allows the virus to infect cells. Prior to this interaction, S protein is needed to be cleaved by different protease enzymes (furins, cathepsins, serine proteases) <ref>DOI 10.1128/AAC.00754-20</ref>. SARS-CoV-2 S protein presents two functional domains S1, the receptor binding domain, and S2, that contains functional elements involved in membrane fusion. There are multiple sites in which this protein can be cleaved; one of these is at the S1/S2 boundary and another within S2. The S1/S2 cleavage site contains multiple arginine residues, which allows the action of serine proteases <ref>DOI 10.1136/jclinpath-2020-206987</ref>. ''Coronaviridae'' family tend to prefer "TMPRSS2" for the cleavage of S protein over other proteases, such as the endosomal cathepsins. As this protease is expressed in SARS-CoV-2 target cells throughout the human respiratory tract, it is also required for the spread of this virus. However, it has been demonstrated to be dispensable for the development or homeostasis of mice models, so it can be considered a potential target to fight the infection of these viruses. | ||
| - | [[Image:CLEAVAGE.jpg|thumb|left|300px|Schematic representation of the cleavage sites in TMPRSS2 protein structure.]] | + | [[Image:CLEAVAGE.jpg|thumb|left|300px|Schematic representation of the cleavage sites in TMPRSS2 protein structure.<ref>DOI 10.1016/j.biochi.2017.07.016</ref>]] |
{{Clear}} | {{Clear}} | ||
| Line 82: | Line 86: | ||
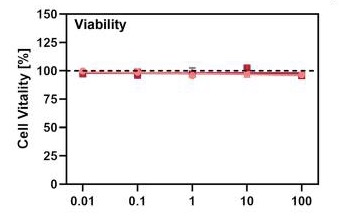
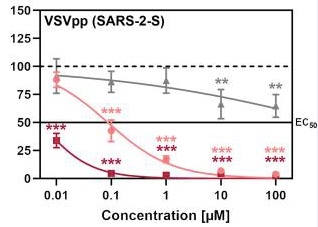
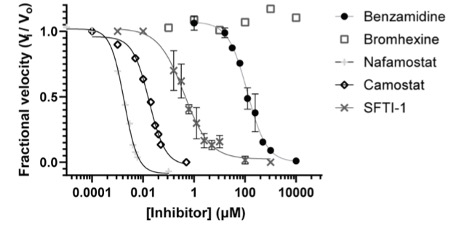
Although nafamostat potently neutralizes TMPRSS2 activity, it is non-selective and disables trypsin-like serine proteases involved in coagulation such as plasmin, FXa, and FXIIa, as well as other TTSPs through its generic arginine-like engagement with the S1 subsite.<ref>DOI 10.1101/2020.06.23.167544</ref><ref>DOI 10.1159/000215139</ref> Also, it requires continuous intravenous infusion to approach therapeutic concentrations for COVID-19 owing to its short biological half-life of 8 minutes. | Although nafamostat potently neutralizes TMPRSS2 activity, it is non-selective and disables trypsin-like serine proteases involved in coagulation such as plasmin, FXa, and FXIIa, as well as other TTSPs through its generic arginine-like engagement with the S1 subsite.<ref>DOI 10.1101/2020.06.23.167544</ref><ref>DOI 10.1159/000215139</ref> Also, it requires continuous intravenous infusion to approach therapeutic concentrations for COVID-19 owing to its short biological half-life of 8 minutes. | ||
| - | [[Image:ACYL.jpg|thumb|left|500px|Close-up view of the SP catalytic triad residues and the post-activation Asp440:Ile256 salt bridge showing complete maturation of the protease. Nafamostat treatment results in phenylguanidino acylation of Ser441. Polar contacts are shown as yellow dashed lines.]] | + | [[Image:ACYL.jpg|thumb|left|500px|Close-up view of the SP catalytic triad residues and the post-activation Asp440:Ile256 salt bridge showing complete maturation of the protease. Nafamostat treatment results in phenylguanidino acylation of Ser441. Polar contacts are shown as yellow dashed lines. <ref>DOI 10.1101/2021.06.23.449282</ref>]] |
{{Clear}} | {{Clear}} | ||
| Line 110: | Line 114: | ||
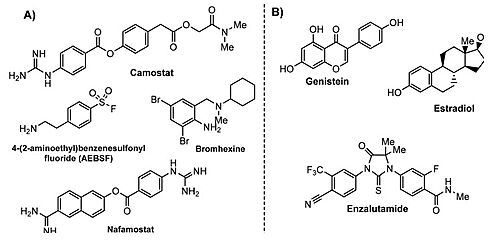
As TMPRSS2 expression seems to be modulated by estrogens and androgens, data suggest that the activation of estrogen pathways or inhibition of androgen pathways may be a new target for therapeutic clinical intervention for symptom amelioration in COVID-19 patients. <ref>DOI 10.20944/preprints202003.0360.v2 | As TMPRSS2 expression seems to be modulated by estrogens and androgens, data suggest that the activation of estrogen pathways or inhibition of androgen pathways may be a new target for therapeutic clinical intervention for symptom amelioration in COVID-19 patients. <ref>DOI 10.20944/preprints202003.0360.v2 | ||
| - | </ref> | + | </ref> In fact, according to this same study estrogen-related compounds and androgen receptor antagonists appear to be the most securely identified down-regulators of TMPRSS2 expression amongst FDA approved drugs and other widely tested compounds. |
| - | + | [[Image:COMPUESTOS.jpg|thumb|left|500px|TMPRSS2 modulators: (A) chemical structure of TMPRSS2 inhibitors and (B) some transcriptional inhibitors. <ref>DOI 10.1021/acs.jmedchem.0c00606</ref>]] | |
| - | + | ||
| - | [[Image:COMPUESTOS.jpg|thumb|left|500px|TMPRSS2 modulators: (A) chemical structure of TMPRSS2 inhibitors and (B) some transcriptional inhibitors.]] | + | |
{{Clear}} | {{Clear}} | ||
| + | ==3D structures of transmembrane protease serine== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[7meq]], [[7xyd]], [[7y0e]] – hTMPRRS2 + drug – human<br /> | ||
| + | [[7y0f]], [[8hd8]] – hTMPRRS2 + inhibitor<br /> | ||
| + | [[2oq5]] – hTMPRRS11e + peptide inhibitor <br /> | ||
| + | [[6kd5]] – hTMPRRS13 + peptide inhibitor <br /> | ||
| + | [[7wqx]] – hTMPRRS15 – Cryo EM <br /> | ||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | </StructureSection> | ||
| + | [[Category:Topic Page]] | ||
Current revision
TMPRSS2 (Transmembrane protease serine 2) is a membrane protein belonging to the type II transmembrane serine protease (TTSP) family. It is functionally classified as a trypsin-like protease (TLP). [1] Serine proteases are known to be involved in many physiological and pathological processes, but the exact function remains unclear. It is involved in two main pathological processes, prostate cancer and viral entry. TMPRSS2 is expressed in several tissues regulated by androgens and undergoes autocatalytic cleavage for its activation.
- Transmembrane protease serine 2 has a role in proteolytic epithelial sodium channel activation[2].
- Transmembrane protease serine 11e is induced during inflammation response[3].
- Transmembrane protease serine 13 facilitates virus entry into host cell by proteolytically cleaving and activating viral envelope glycoproteins.
- Transmembrane protease serine 15 converts pancreatic trypsinogen to trypsin.
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Paula R. Mallavibarrena, Ines Muniesa-Martinez, Michal Harel, Laura Aleixos Juan, Jaime Prilusky