We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1697
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
== Biological relevance and broader implications == | == Biological relevance and broader implications == | ||
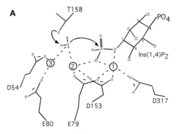
| - | This protein is involved in dephosphorylation of inositol which inhibits inositol signaling in cattle. The two metal ions, Magnesium and Lithium, are particularly important in activation and inhibition of inositol signaling. Since these enzymes are seen in many different organisms, with slight mutations in the motif of the enzyme without change in function, understanding the mechanics of enzyme inhibition in this metabolic pathway would allow a better understanding of metabolic pathways. | + | This protein is involved in dephosphorylation of inositol which inhibits inositol signaling in cattle. The metal ions assist in holding the water so that it can attack the phosphate to take it off. The two metal ions, Magnesium and Lithium, are particularly important in activation and inhibition of inositol signaling. Since these enzymes are seen in many different organisms, with slight mutations in the motif of the enzyme without change in function, understanding the mechanics of enzyme inhibition in this metabolic pathway would allow a better understanding of metabolic pathways. |
== Important amino acids== | == Important amino acids== | ||
Current revision
| This Sandbox is Reserved from 10/01/2021 through 01/01//2022 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1690 through Sandbox Reserved 1699. |
To get started:
More help: Help:Editing |
7KIR
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Dollins DE, Xiong JP, Endo-Streeter S, Anderson DE, Bansal VS, Ponder JW, Ren Y, York JD. A Structural Basis for Lithium and Substrate Binding of an Inositide Phosphatase. J Biol Chem. 2020 Nov 10. pii: RA120.014057. doi: 10.1074/jbc.RA120.014057. PMID:33172890 doi:http://dx.doi.org/10.1074/jbc.RA120.014057