Malarial Dihydrofolate Reductase as Drug Target
From Proteopedia
| Line 25: | Line 25: | ||
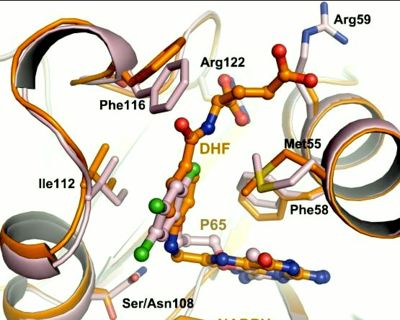
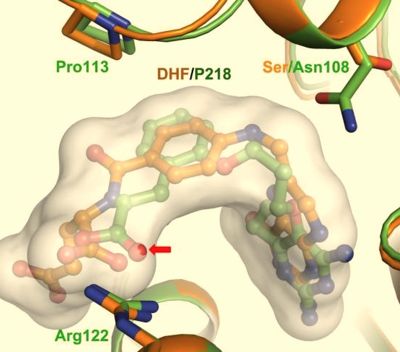
It was resolved that a structure containing 2,4-diaminopyrimidine anchor, such as that of P65, on a new drug would provide a solution for the steric hindrance present with pyrimethamine because of its rigid chlorophenyl group. This anchor allows deep binding into the active site of PfDHFR. On the other end of the molecule connected by a flexible, five-atom linker region, there would be a carboxyl group, that would form strong hydrogen bonds with the conserved <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Pfdhfr_arg122/1'>Arg122</scene> residue. The flexible linker would reduce the chance of resistance from mutations because of its decreased specificity that can occur due to the strong anchor and other end with hydrogen bonds. These ideas culminated into the formation of P218. | It was resolved that a structure containing 2,4-diaminopyrimidine anchor, such as that of P65, on a new drug would provide a solution for the steric hindrance present with pyrimethamine because of its rigid chlorophenyl group. This anchor allows deep binding into the active site of PfDHFR. On the other end of the molecule connected by a flexible, five-atom linker region, there would be a carboxyl group, that would form strong hydrogen bonds with the conserved <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Pfdhfr_arg122/1'>Arg122</scene> residue. The flexible linker would reduce the chance of resistance from mutations because of its decreased specificity that can occur due to the strong anchor and other end with hydrogen bonds. These ideas culminated into the formation of P218. | ||
| - | There are three regions of the DHFR active site around the conserved arginine that differ between <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Human_dhfr_and_folate/1'>human</scene> and ''Plasmodium falciparum'' that allow a drug to target the PfDHFR specifically while not harming that of humans. The Met55, Cys/Arg59, and Phe116 residues in the PfDHFR are replaced by Phe31, Gln35, and Asn64 at structurally equivalent positions in human DHFR which are crucial differentiations that can be taken advantage of by inhibitors, especially with a carboxyl group, to control species specificity. The plan in devising a compound, such as P218, that would interact with the region surrounding the conserved arginine residue mentioned above is that the compound would interact with the <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Pfdhfr_arg122_region/2'>PfDHFR Arg122 residue region</scene> while leaving the <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Hdhfr_arg70_region/1'>human Arg70 region</scene> and thus human DHFR alone. This design and intent for P218 was shown to be successful. In the quadruple mutant of PfDHFR, the P218 carboxylate side chain makes two hydrogen bonds with the Arg122 residue while in human DHFR, it has no interaction with the Arg70 residue. In the human active site, the P218 side chain to folds back toward its pyrimidine, which pushes Phe31 into a position toward the back of the substrate binding site further preventing the side-chain hydrogen bond formation with Arg70.<ref | + | There are three regions of the DHFR active site around the conserved arginine that differ between <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Human_dhfr_and_folate/1'>human</scene> and ''Plasmodium falciparum'' that allow a drug to target the PfDHFR specifically while not harming that of humans. The Met55, Cys/Arg59, and Phe116 residues in the PfDHFR are replaced by Phe31, Gln35, and Asn64 at structurally equivalent positions in human DHFR which are crucial differentiations that can be taken advantage of by inhibitors, especially with a carboxyl group, to control species specificity. The plan in devising a compound, such as P218, that would interact with the region surrounding the conserved arginine residue mentioned above is that the compound would interact with the <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Pfdhfr_arg122_region/2'>PfDHFR Arg122 residue region</scene> while leaving the <scene name='Malarial_Dihydrofolate_Reductase_as_Drug_Target/Hdhfr_arg70_region/1'>human Arg70 region</scene> and thus human DHFR alone. This design and intent for P218 was shown to be successful. In the quadruple mutant of PfDHFR, the P218 carboxylate side chain makes two hydrogen bonds with the Arg122 residue while in human DHFR, it has no interaction with the Arg70 residue. In the human active site, the P218 side chain to folds back toward its pyrimidine, which pushes Phe31 into a position toward the back of the substrate binding site further preventing the side-chain hydrogen bond formation with Arg70.<ref name="yu" /> The changes in residues around Arg70 sterically prevent P218 from interacting with the residue like it does in PfDHFR. |
Current revision
Introduction
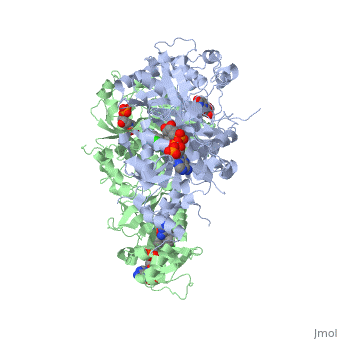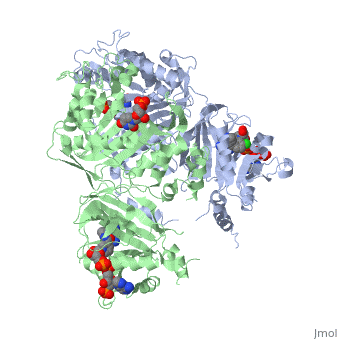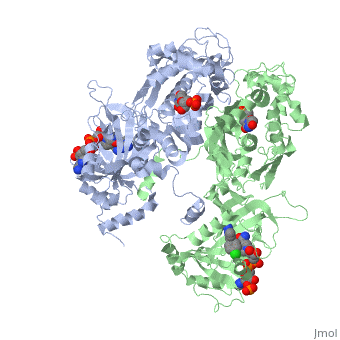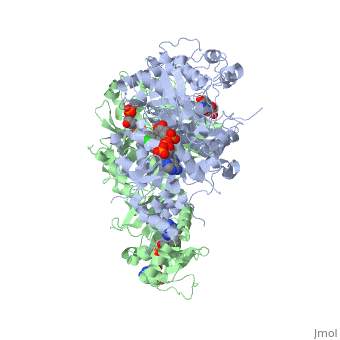
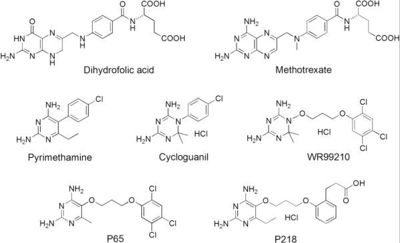
Dihydrofolate reductase (DHFR) plays an essential role in the formation of DNA by managing folate, an organic molecule that shuttles carbons to enzymes that need them for their reactions. Relevant to this context of DHFR involvement, folate gives its carbons to thymidylate synthase, which then uses these carbons to make thymine bases. After folate has removed its carbon atoms, it is the job of DHFR to recycle it. It does this by transferring hydrogen atoms from NADPH to folate to restore it to its useful, reduced form.[1] DHFR is used in all organisms however, each organism makes a lsightly different version. The malaria version of the enzyme has shown to be reliable and the best target for antimalarial drugs. The current antimalarial drugs that target the malarial DHFR include pyrimethamine and cycloguanil. Pyrimethamine is currently used in combination sulfadoxine, a sulfadrug, to treat and prevent malaria but can result in serious side effects including liver damage.[2][3] Recently the effectiveness of these drugs has decreased because of mutations in the enzyme that have led to drug resistance. Since these mutations are becoming much more prevalent in malaria cases, new research in drug development must now incorporate both the wild-type as well as the quadruple mutant DHFR from the Plasmodium falciparum malarial strain, the most common and lethal of the malaria species.[4]
| |||||||||||
References
- ↑ Goodsell, David. "Dihydrofolate Reductase." RCSB PDB-101. RCSB PDB, Oct. 2002. Web. <http://www.rcsb.org/pdb/101/motm.do?momID=34>.
- ↑ "Pyrimethamine & Sulfadoxine." United States National Library of Medicine, n.d. Web. <http://livertox.nih.gov/PyrimethamineSulfadoxine.htm>.
- ↑ "Pyrimethamine and Sulfadoxine (Oral Route)." Mayo Clinic. Mayo Foundation for Medical Education and Research, 01 Nov. 2011. Web. <http://www.mayoclinic.com/health/drug-information/DR600357>.
- ↑ Somsak V, Uthaipibull C, Prommana P, Srichairatanakool S, Yuthavong Y, Kamchonwongpaisan S. Transgenic Plasmodium parasites stably expressing Plasmodium vivax dihydrofolate reductase-thymidylate synthase as in vitro and in vivo models for antifolate screening. Malar J. 2011 Oct 7;10:291. PMID: 21981896
- ↑ Huang F, Tang L, Yang H, Zhou S, Liu H, Li J, Guo S. Molecular epidemiology of drug resistance markers of Plasmodium falciparum in Yunnan Province, China. Malar J. 2012 Jul 28;11:243. PMID: 22839209
- ↑ 6.00 6.01 6.02 6.03 6.04 6.05 6.06 6.07 6.08 6.09 6.10 6.11 Yuthavong Y, Tarnchompoo B, Vilaivan T, Chitnumsub P, Kamchonwongpaisan S, Charman SA, McLennan DN, White KL, Vivas L, Bongard E, Thongphanchang C, Taweechai S, Vanichtanankul J, Rattanajak R, Arwon U, Fantauzzi P, Yuvaniyama J, Charman WN, Matthews D. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16823-8. doi:, 10.1073/pnas.1204556109. Epub 2012 Oct 3. PMID:23035243 doi:http://dx.doi.org/10.1073/pnas.1204556109
Proteopedia Page Contributors and Editors (what is this?)
Mary Smith, Alexander Berchansky, Karsten Theis, Michal Harel