We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Test160122
From Proteopedia
(Difference between revisions)
| (35 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='3i3f' size='350' side='right' scene='90/901530/Cv/ | + | <StructureSection load='3i3f' size='350' side='right' scene='90/901530/Cv/14' caption=''> |
| - | + | '''23/01/22''' | |
| - | *<scene name='90/901530/Cv/11'>Surface</scene> | + | *<scene name='90/901530/Cv/24'>Tusual, color is chosen same as atom after clicking on atom</scene> - after that, the label color is remaining white |
| + | *Everything is OK: <scene name='90/901530/Cv/30'>Similar to Tusual, color is chosen same as atom before clicking on atom</scene> | ||
| + | |||
| + | *<scene name='90/901530/Cv/26'>Trot</scene> | ||
| + | *<scene name='90/901530/Cv/27'>Trock</scene> | ||
| + | *<scene name='90/901530/Cv/28'>Twobble</scene> | ||
| + | |||
| + | |||
| + | *<scene name='90/901530/Cv1/1'>Surfacenew</scene> | ||
| + | *<scene name='90/901530/Cv1/2'>Surfacenew1</scene> | ||
| + | *<scene name='90/901530/Cv1/3'>Morph_Loop_6models</scene><jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Stop Animation</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | *<scene name='90/901530/Cv1/4'>Morph_Palindrom_2models</scene><jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Stop Animation</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | |||
| + | *<scene name='90/901530/L22s/2'>Mutation L22S (copy saved in present page): </scene> | ||
| + | <jmol> | ||
| + | <jmolLink> | ||
| + | <script> | ||
| + | animation on; animation mode loop; frame 1 1 play; animation off; frame 1; select [LEU]22;color selectionHalos green;selectionHalos on; | ||
| + | </script> | ||
| + | <text>Wild type </text> | ||
| + | </jmolLink> | ||
| + | </jmol> | ||
| + | conformation and | ||
| + | <jmol> | ||
| + | <jmolLink> | ||
| + | <script> | ||
| + | animation on; animation mode loop; frame 2 2 play; animation off; frame 2; select [SER]22;color selectionHalos red;selectionHalos on; | ||
| + | </script> | ||
| + | <text>mutation. | ||
| + | </text> | ||
| + | </jmolLink> | ||
| + | </jmol> | ||
| + | Click here to see the | ||
| + | <jmol> | ||
| + | <jmolLink> | ||
| + | <script> | ||
| + | animation off; frame all; select [SER]22;color selectionHalos red;selectionHalos on; | ||
| + | </script> | ||
| + | <text>mutated and wild type | ||
| + | </text> | ||
| + | </jmolLink> | ||
| + | </jmol> residues together, and an | ||
| + | <jmol> | ||
| + | <jmolLink> | ||
| + | <script> | ||
| + | animation on; animation mode loop; frame play; select [SER]22;color selectionHalos red;selectionHalos on; | ||
| + | </script> | ||
| + | <text>animation | ||
| + | </text> | ||
| + | </jmolLink> | ||
| + | </jmol> between the two states. Putative new hydrogen bonds were added. | ||
| + | <jmol><jmolButton> | ||
| + | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
| + | <text>Stop Animation</text> | ||
| + | </jmolButton></jmol> | ||
| + | |||
| + | *<scene name='90/901530/Cv1/5'>From uploaded file</scene> | ||
| + | |||
| + | |||
| + | |||
| + | --------- | ||
| + | *<scene name='90/901530/Cv/4'>160122was_with_labels/now180122_without_labels</scene> | ||
| + | |||
| + | *<scene name='90/901530/Cv/16'>180122saved_today_with_labels/in_text_without_labels</scene> | ||
| + | *<scene name='90/901530/Cv/17'>180122saved_today/should_be_simple_copy_of_previous_scene_without_rotation</scene> | ||
| + | *<scene name='90/901530/Cv/11'>Surface</scene> | ||
| + | *<scene name='90/901530/Cv/19'>Te</scene> | ||
| + | *<scene name='90/901530/Cv/23'>Terot</scene> | ||
| + | *<scene name='90/901530/Cv/22'>Tewobble</scene> | ||
*<scene name='90/901530/Cv/3'>Test1</scene> | *<scene name='90/901530/Cv/3'>Test1</scene> | ||
| - | + | ||
*<scene name='90/901530/Cv/5'>Te3</scene> | *<scene name='90/901530/Cv/5'>Te3</scene> | ||
*<scene name='90/901530/Cv/10'>T8</scene> <jmol><jmolButton> | *<scene name='90/901530/Cv/10'>T8</scene> <jmol><jmolButton> | ||
| Line 11: | Line 87: | ||
<text>Stop Animation</text> | <text>Stop Animation</text> | ||
</jmolButton></jmol> | </jmolButton></jmol> | ||
| - | *<scene name='90/901530/Cv/ | + | *<scene name='90/901530/Cv/12'>T7</scene> <jmol><jmolButton> |
<script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | <script>if (_animating); anim pause;set echo bottom left; color echo white; font echo 20 sansserif;echo Animation Paused; else; anim resume; set echo off;endif;</script> | ||
<text>Stop Animation</text> | <text>Stop Animation</text> | ||
| Line 62: | Line 138: | ||
</jmolButton></jmol> | </jmolButton></jmol> | ||
| - | [[Image: | + | ------------------------------ |
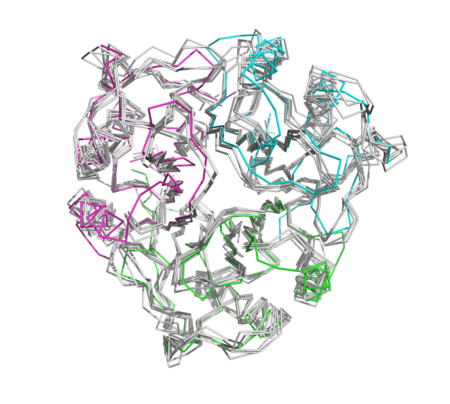
| + | [[Image:Figure3newt.png|left|450px|thumb|NEW: Despite the low sequence similarity the hypothetical protein is a prototypical YjgF/YER057c/UK114 endoribonuclease]] | ||
{{Clear}} | {{Clear}} | ||
| - | [[Image: | + | [[Image:Trimercomparison1.png|left|450px|thumb|NEW]] |
{{Clear}} | {{Clear}} | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| |||||||||||