Sandbox Reserved 1660
From Proteopedia
(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 26: | Line 26: | ||
<p align="justify"> | <p align="justify"> | ||
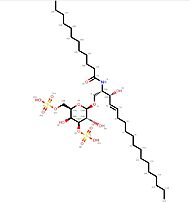
| - | The ligands that can be presented by CD1d to NKT or other CD1d-restricted T cells are quite specific, because the recognition is based on a hydrophobic interaction between the ligand and the CD1d molecule. Thus, it limits the risk | + | The ligands that can be presented by CD1d to NKT or other CD1d-restricted T cells are quite specific, because the recognition is based on a hydrophobic interaction between the ligand and the CD1d molecule. Thus, it limits the risk for other similar molecules to bind CD1d proteins and ensures that the immune response is accurate. Among these CD1d ligands, glycolipids from a marine sponge (alphagalactosylceramide or α-GalCer), bacterial glycolipids, normal endogenous glycolipids, tumor-derived phospholipids and glycolipids, and nonlipidic molecules have been described<ref name="Immunity">Brutkiewicz, R. R. CD1d Ligands: The Good, the Bad, and the Ugly. The Journal of Immunology 2006, 177 (2), 769–775. https://doi.org/10.4049/jimmunol.177.2.769.</ref>. |
</p> | </p> | ||
<br> | <br> | ||
| Line 38: | Line 38: | ||
The alpha chain is made of three domains: alpha 1, alpha 2 and alpha 3. The association of alpha 1 and alpha 2 is composed of two [https://en.wikipedia.org/wiki/Beta_sheet beta-sheets] and a set of 2 [https://proteopedia.org/wiki/index.php/Alpha_helix alpha helixes]. Each beta-sheet contains four antiparallel strands. The ligand binds between the two alpha 1 and 2 helices. The alpha 3 domain is non-covalently bound with beta-2-microglobulin domain. | The alpha chain is made of three domains: alpha 1, alpha 2 and alpha 3. The association of alpha 1 and alpha 2 is composed of two [https://en.wikipedia.org/wiki/Beta_sheet beta-sheets] and a set of 2 [https://proteopedia.org/wiki/index.php/Alpha_helix alpha helixes]. Each beta-sheet contains four antiparallel strands. The ligand binds between the two alpha 1 and 2 helices. The alpha 3 domain is non-covalently bound with beta-2-microglobulin domain. | ||
Additionally, there are five <scene name='86/868193/Oligosaccharides/4'>oligosaccharides</scene><ref name="Structure"/> bound to the alpha chain via N-glycosylations, three of which have been clearly identified<ref name="oligo">Sriram, V., Willard, C.A., Liu, J., & Brutkiewicz, R.R.(2008). Importance of N-linked glycosylation in the functional expression of murine CD1d1. Immunology, 123:272–281.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2433293/</ref>. The total molecular weight of the alpha chain is 33 kDa when not associated to any oligosaccharide and 55 kDa when oligosaccharides are associated to the chain<ref name="properties"/>.</p> | Additionally, there are five <scene name='86/868193/Oligosaccharides/4'>oligosaccharides</scene><ref name="Structure"/> bound to the alpha chain via N-glycosylations, three of which have been clearly identified<ref name="oligo">Sriram, V., Willard, C.A., Liu, J., & Brutkiewicz, R.R.(2008). Importance of N-linked glycosylation in the functional expression of murine CD1d1. Immunology, 123:272–281.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2433293/</ref>. The total molecular weight of the alpha chain is 33 kDa when not associated to any oligosaccharide and 55 kDa when oligosaccharides are associated to the chain<ref name="properties"/>.</p> | ||
| - | CD1d molecules are structurally similar to [[Major Histocompatibility Complex Class I]], but present lipid antigens as opposed to peptides. Thus the cleft where the ligand can bind is different between MHC molecules and CD1d molecules. Indeed, the hydrophobic cleft of CD1d has a narrow opening.The recognition between the protein and its ligand occurs at a specific hydrophobic spot which creates an appropriate environment for the interaction to happen. This <scene name='86/868193/Site/ | + | CD1d molecules are structurally similar to [[Major Histocompatibility Complex Class I]], but present lipid antigens as opposed to peptides. Thus the cleft where the ligand can bind is different between MHC molecules and CD1d molecules. Indeed, the hydrophobic cleft of CD1d has a narrow opening.The recognition between the protein and its ligand occurs at a specific hydrophobic spot which creates an appropriate environment for the interaction to happen. This <scene name='86/868193/Site/7'>site</scene> is located at the A' and F' pockets in the region of the alpha helices<ref name="site">Schiefner, A.; Fujio, M.; Wu, D.; Wong, C.-H.; Wilson, I. A. Structural Evaluation of Potent NKT-Cell Agonists: Implications for Design of Novel Stimulatory Ligands. J Mol Biol 2009, 394 (1), 71–82. https://doi.org/10.1016/j.jmb.2009.08.061</ref>. |
<br><br> | <br><br> | ||
=='''Impact of ligand-binding'''== | =='''Impact of ligand-binding'''== | ||
| Line 58: | Line 58: | ||
<p align="justify"> | <p align="justify"> | ||
| - | The presentation of several | + | The presentation of several kinds of ligands can have immunopotentiating effects, such as serving as an adjuvant in [https://en.wikipedia.org/wiki/Malaria malaria] vaccine or resulting in a more rapid clearance of certain [https://en.wikipedia.org/wiki/Virus virus] infections. They can also be protective in autoimmune diseases or cancer<ref name="affinity"/><ref name="Immunity"/><ref>Sköld, M.; Behar, S. M. Role of CD1d-Restricted NKT Cells in Microbial Immunity. Infect Immun 2003, 71 (10), 5447–5455. https://doi.org/10.1128/IAI.71.10.5447-5455.2003.</ref>. |
</p> | </p> | ||
Current revision
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
| |||||||||||