SARS-CoV-2 protein M
From Proteopedia
(Difference between revisions)
| (16 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='' size='340' side='right' caption='Caption for this structure' scene='84/842059/Sars-cov-2_protein_m/2'> | |
| - | <StructureSection load='' size='340' side='right' caption='Caption for this structure' scene='84/842059/Sars-cov-2_protein_m/ | + | |
{{Theoretical_model}} {{Template:ModelConfidence}} | {{Theoretical_model}} {{Template:ModelConfidence}} | ||
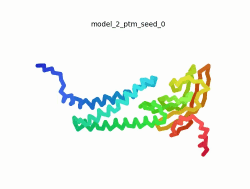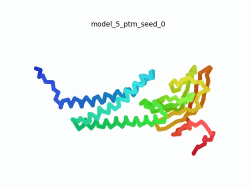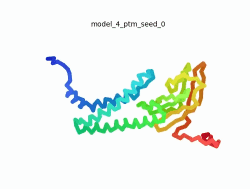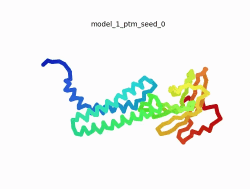
| - | To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein | + | To the right is an '''[[AlphaFold]]2''' 3D model of SARS CoV-2 Protein M (length=222 amino acids, [https://www.uniprot.org/uniprot/P0DTC5 UniProt ID: P0DTC5]) color coded by the pLDDT scores. It corresponds to the highest ranked model in terms of the pLDDT confidence scores, ''i.e.'', model 4<ref name="MIT_ColabFold">[https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/beta/AlphaFold2_advanced.ipynb MIT ColabFold]</ref>. |
| + | |||
== Function == | == Function == | ||
| Line 9: | Line 9: | ||
== Function == | == Function == | ||
'''Protein M''' | '''Protein M''' | ||
| - | Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins.<ref>[https://zhanglab.ccmb.med.umich.edu/COVID-19/ Modeling of the SARS-COV-2 Genome]</ref><ref>pmid 32200634</ref> | + | Component of the viral envelope that plays a central role in virus morphogenesis and assembly via its interactions with other viral proteins.<ref name="MIT_ColabFold"/><ref name="Zhang_Lab">[https://zhanglab.ccmb.med.umich.edu/COVID-19/ Modeling of the SARS-COV-2 Genome]</ref> |
| + | <ref>pmid 32200634</ref>. | ||
== Disease == | == Disease == | ||
| Line 16: | Line 17: | ||
== Structural highlights == | == Structural highlights == | ||
| - | [[Image:SARSCoV2_protein_M_250.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein M''', ''rainbow'' color coded N-C<ref name="MIT_ColabFold"/>.]] | + | [[Image:SARSCoV2_protein_M_250.gif|250px|left|thumb|'''Morph''' of the top 5 ranked [[AlphaFold]]2 models of '''SARS-CoV-2 Protein M''', ''rainbow'' color coded N-C (blue to red)<ref name="MIT_ColabFold"/>.]] |
| Line 40: | Line 41: | ||
== See also == | == See also == | ||
| - | [[Coronavirus_Disease 2019 (COVID-19)]] | + | [[Coronavirus_Disease 2019 (COVID-19)]]<br> |
| + | [[SARS-CoV-2_virus_proteins]]<br> | ||
| + | [[COVID-19 AlphaFold2 Models]] | ||
__NOTOC__ | __NOTOC__ | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 MIT ColabFold
- ↑ Modeling of the SARS-COV-2 Genome
- ↑ Zhang C, Zheng W, Huang X, Bell EW, Zhou X, Zhang Y. Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J Proteome Res. 2020 Apr 3;19(4):1351-1360. doi: 10.1021/acs.jproteome.0c00129., Epub 2020 Mar 24. PMID:32200634 doi:http://dx.doi.org/10.1021/acs.jproteome.0c00129