We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
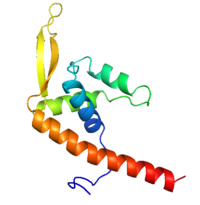
Replication Termination Protein
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 119: | Line 119: | ||
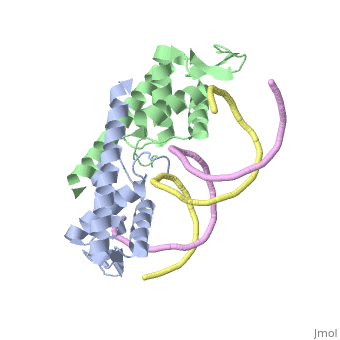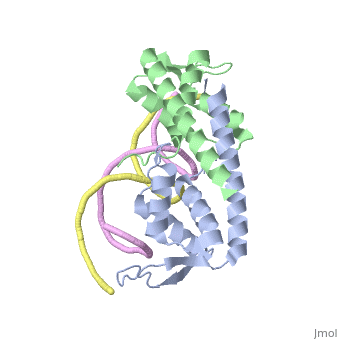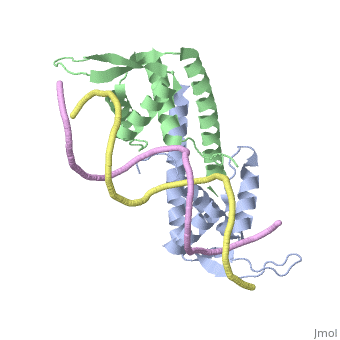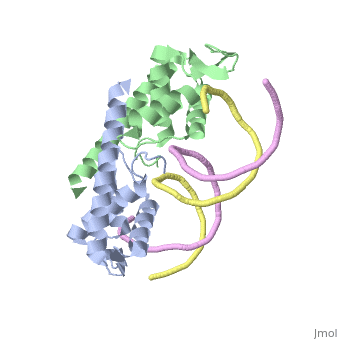
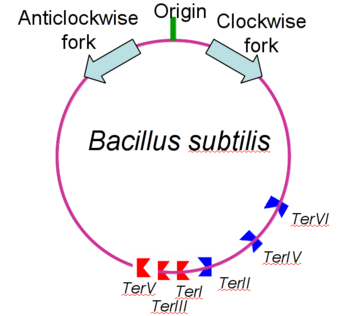
Debate over the mechanism of Replication termination by RTP has led to a number of mechanisms being proposed. Original work on this question was hampered by an incorrect structure of RTP being released. This structure was produced in solution and was shown to be a symmetric dimer<ref> Bastia D. (1995) Crystal structure of the replication terminator protein from b. subtilis at 2.6 A. Cell 80: 651-660 </ref> Later crystallisation of the protein bound to DNA showed the RTP formed a asymmetric dimer when bound to cognate sequences <ref> Wilce et. al. (2001) Structure of the RTP−DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology, 8:206-210</ref> . One early study proposed that the asymmetric interaction between terminator protein and terminator DNA contributed to the observed polarity, with later studies showing that the RTP/Ter complex contained partially unwound DNA, suggesting a locked complex was responsible. More recent data has implicated that protein -protein interactions between RTP and the helicase is primarily responsible for termination and that this mechanism is modulated by the asymmetrical interactions described above <ref>Kaplan D.L., Bastia D. (2009). Mechanisms of polar arrest of a replication fork. Molecular Microbiology 72: 279-284</ref>. | Debate over the mechanism of Replication termination by RTP has led to a number of mechanisms being proposed. Original work on this question was hampered by an incorrect structure of RTP being released. This structure was produced in solution and was shown to be a symmetric dimer<ref> Bastia D. (1995) Crystal structure of the replication terminator protein from b. subtilis at 2.6 A. Cell 80: 651-660 </ref> Later crystallisation of the protein bound to DNA showed the RTP formed a asymmetric dimer when bound to cognate sequences <ref> Wilce et. al. (2001) Structure of the RTP−DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology, 8:206-210</ref> . One early study proposed that the asymmetric interaction between terminator protein and terminator DNA contributed to the observed polarity, with later studies showing that the RTP/Ter complex contained partially unwound DNA, suggesting a locked complex was responsible. More recent data has implicated that protein -protein interactions between RTP and the helicase is primarily responsible for termination and that this mechanism is modulated by the asymmetrical interactions described above <ref>Kaplan D.L., Bastia D. (2009). Mechanisms of polar arrest of a replication fork. Molecular Microbiology 72: 279-284</ref>. | ||
| - | + | ||
==3D structures of replication termination protein== | ==3D structures of replication termination protein== | ||
| - | + | [[Replication termination protein 3D structures]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==References== | ==References== | ||
{{reflist}} | {{reflist}} | ||
| - | + | </StructureSection> | |
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Craig Mooney, Joel L. Sussman