We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1705
From Proteopedia
(Difference between revisions)
| (44 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Template:CH462_Biochemistry_II_2022}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Template:CH462_Biochemistry_II_2022}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | <StructureSection load='7n00' size='340' side='right' caption='Anaplastic Lymphoma Kinase' PDB:[https://www.rcsb.org/structure/7N00]' scene='90/904310/Dimer_ligand_complex/ | + | <StructureSection load='7n00' size='340' side='right' caption='Anaplastic Lymphoma Kinase' PDB:[https://www.rcsb.org/structure/7N00]' scene='90/904310/Dimer_ligand_complex/5'> |
= Anaplastic Lymphoma Kinase (ALK) = | = Anaplastic Lymphoma Kinase (ALK) = | ||
== Introduction == | == Introduction == | ||
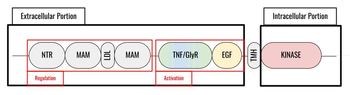
| + | [[Image:ALK_Domains.png|350 px|right|thumb|Figure 1. Outline of the domains and regions of anaplastic lymphoma kinase]] | ||
| + | Anaplastic lymphoma kinase is a [https://en.wikipedia.org/wiki/Receptor_tyrosine_kinase receptor tyrosine kinase] (RTK) important to the regulation of functions within the central nervous system <ref name="Reshetnyak">PMID:34819673</ref>. RTKs are the high-affinity cell surface receptors for many polypeptide growth factors, cytokines, and hormones. ALK activates pathways that promote cell growth and proliferation, similar to the Insulin Receptor (IR). These pathways include, but are not limited to, the [https://en.wikipedia.org/wiki/MAPK/ERK_pathway ERK], [https://en.wikipedia.org/wiki/JAK-STAT_signaling_pathway JAK], and [https://en.wikipedia.org/wiki/PI3K/AKT/mTOR_pathway PI3K] pathways. ALK is composed of two identical monomers consisting of seven unique domains, and two intermixed regions. Among these, one of the most notable is the glycine-rich region (GlyR), characterized by helical structures formed by glycine residues. Other domains are archetypal of RTK activation and regulation. Several potential human ALK ligands have been discovered, however two ALK activating ligands (ALKALs) have been further studied for their ability to activate ALK, ALK homologs, and ALK fusion proteins <ref name="Huang">PMID: 30400214</ref>. The preferred ALKAL for activation of the kinase domain is AUG, which binds in a dimeric fashion to ALK, inducing a conformational change. In order to determine the atomic details of human ALK dimerization and activation by AUG, the methods of [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryo-electron microscopy], [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance_spectroscopy nuclear magnetic resonance], and [https://en.wikipedia.org/wiki/X-ray_crystallography X-ray crystallography] were utilized <ref name="Reshetnyak">PMID:34819673</ref>. Due to its role in cell growth and proliferation signaling, ALK is considered to be a proto-oncogene, meaning that it is capable of producing mutations that lead to cancer. ALK mutations can lead to various types of cancers, including non-small-cell lung cancer, anaplastic large cell lymphoma, squamous cell carcinoma, and inflammatory myofibroblastic cancer <ref name="Palmer">PMID:19459784</ref>. | ||
| + | |||
| - | Anaplastic lymphoma kinase is a receptor tyrosine kinase (RTK) that is important in regulating functions within the central nervous system <ref name="Reshetnyak">PMID:34819673</ref>. The route to discovery of this protein's structure was rather complex, spanning almost 20 years; the kinase domain was discovered in 1994, the full protein structure in 1997, and the ligand structures in 2014. These structures were found using [https://www.chemistryworld.com/news/explainer-what-is-cryo-electron-microscopy/3008091.article cryo-electron microscopy],[https://www.britannica.com/science/nuclear-magnetic-resonance nuclear magnetic resonance], and [https://www.sciencemuseum.org.uk/objects-and-stories/chemistry/x-ray-crystallography-revealing-our-molecular-world#:~:text=X%2Dray%20crystallography%20is%20a,called%20an%20x%2Dray%20camera. X-ray crystallography]. Anaplastic lymphoma kinase is a proto-oncogene with mutations associated with various types of cancers, including non-small-cell lung cancer, anaplastic large cell lymphoma, squamous cell carcinoma, and inflammatory myofibroblastic cancer <ref name="Palmer">PMID:19459784</ref>. | ||
== General Structure == | == General Structure == | ||
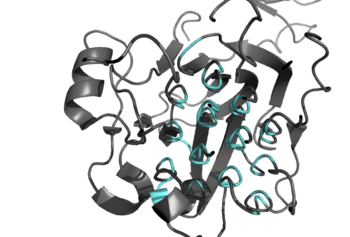
| - | Anaplastic lymphoma kinase is a <scene name='90/904310/Dimer/ | + | [[Image:Ray_Traced_GlyR_Image.png|350 px|left|thumb|Figure 2: Shown in teal is the Glycine Rich Region of ALK in its helical structure. ]] |
| + | Anaplastic lymphoma kinase is a <scene name='90/904310/Dimer/5'>homodimer</scene>, in which each <scene name='90/904310/Monomer_a/3'>monomer</scene> contains seven domains and two regions <ref name="Tongqing">PMID:34819665</ref>. These domains and regions are as follows: N-terminal region (NTR), two meprin–A-5 protein–receptor protein tyrosine phosphatase μ domains (MAM), low density lipoprotein receptor class A domain (LDL), <scene name='90/904310/Tnf_highlighted_monomer/5'>tumor necrosis factor receptor-like domain</scene> (TNF), <scene name='90/904310/Glyr_highlighted_monomer/5'>glycine rich region</scene> (GlyR), <scene name='90/904310/Egf_highlighted_monomer/3'>epidermal growth factor receptor-like domain</scene> (EGF), transmembrane α-helix (TMH), kinase domain <ref name="Reshetnyak">PMID:34819673</ref>. The NTR functions as a signal peptide, the structure of which is yet to be determined. Both MAM domains and the LDL domain are unique to ALK within the superfamily of RTKs, and the biological role of these domains has not yet been determined <ref name="Huang">PMID: 30400214</ref>. However, the LDL and MAM regions are thought to be involved in various regulatory functions; studies of other LDL protein domains indicate a role for LDL domains in plasma cholesterol homeostasis <ref name="Pedersen">PMID:24798328</ref>. Additionally, research on MAM domains implicates a role in cell-cell interactions through homophilic binding <ref name="Huang">PMID: 30400214</ref>. The TNF-like domain assists in mediating mature T-cell receptor induced apoptosis. Both the TNF domain and GlyR region are discontinuous, traversing each other frequently <ref name="Reshetnyak">PMID:34819673</ref>. The GlyR region consists of several glycine helices, a highly unique structure. Though the function of ALK's EGF domain is unknown, EGF domains are typically found in the extracellular region and are thought to be important building blocks for extracellular proteins <ref name="Selander-Sunnerhagen">PMID:1527084</ref>. The TMH connects the extracellular and intracellular regions of ALK through the plasma membrane. The kinase domain is in the intracellular region and is autoactivated through phosphorylation at positions <scene name='90/904309/Tyrosines/1'>Y1278, Y1282, and Y1283</scene>. This allows the RTK to begin signaling cascades throughout the cell <ref name="Selander-Sunnerhagen">PMID:1527084</ref>. While the structures of the N-terminal region, MAM, and LDL have not been determined, it has been shown that only the TNF, GlyR, and EGF portions of ALK are required in the extracellular region for ligand activation via ALKALs <ref name="Palmer">PMID: 19459784</ref>. All portions of anaplastic lymphoma kinase are located in the extracellular domain except for the transmembrane α-helix, which is in the transmembrane region, and the kinase domain located in the intracellular region. | ||
| + | |||
=== Ligand Binding=== | === Ligand Binding=== | ||
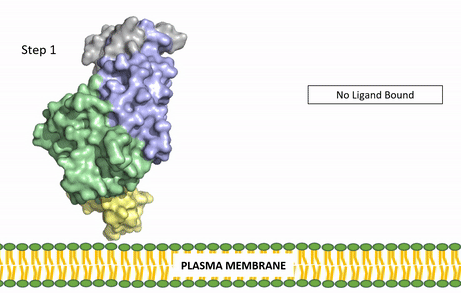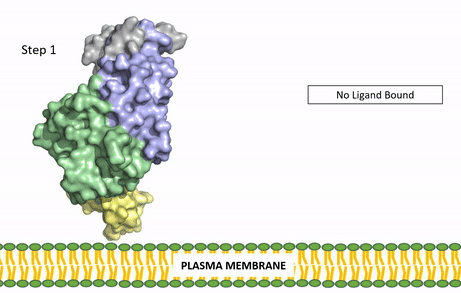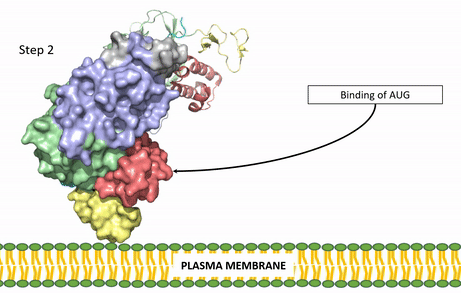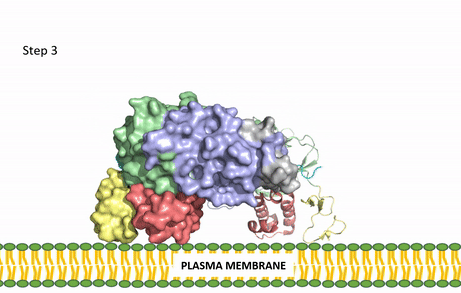
| - | The | + | [[Image:ALK Conformational Change Gif.gif|850 px|right|thumb|Figure 3: Gif-image of the conformational change occurring in the extracellular region of Anaplastic Lymphoma Kinase once the AUG ligand has bound to the ligand-binding site. This change is stabilized through contacts of the AUG and the plasma membrane. The video was made using stop motion animation techniques, then converted to gif format using EZgif.]] |
| + | ALKALs recognized by ALK are FAM150, binding in either a monomeric or dimeric fashion, and <scene name='90/904310/Ligand/2'>AUG</scene> in a dimeric fashion <ref name="Li">PMID:34819665</ref> . Its biologically preferred ligand is AUG, a protein ligand containing 128 residues that form three α-helices. FAM150 is a structural ortholog of AUG. The binding of ALK to it's ligand results in homodimerization and an induced conformational change. Prior to the ligand binding to ALK, the extracellular domain is oriented vertically and perpendicularly to the plasma membrane (Step 1, Figure 3). Once the ligand is <scene name='90/904310/Dimer_ligand_complex/5'>bound</scene> (Step 2, Figure 3), ALK undergoes a conformational change and folds over so that the <scene name='90/904310/Positive_residues/1'>positively charged residues</scene> on the ALKAL can now interact with the negatively charged phosphate heads of the plasma membrane (Step 3, Figure 3). The residues of ALK and its ligand interact through the formation of <scene name='90/904310/Dimer-ligand-interface/5'>salt bridges</scene><ref name="Munck">PMID:34646012</ref>. It has been hypothesized that the GlyR region plays a role in the flexibility needed to complete the conformational change. This conformational change via ligand activation allows for the auto-activation of the kinase domain, where the domains use the tyrosine phosphorylation mechanism to phosphorylate tyrosine residues on the opposite monomer. ALK is a Class II RTK ([[Insulin Receptor|Insulin Receptor (IR)]] Family), sharing much of its kinase activation mechanism with the IR. | ||
| + | |||
| + | |||
| + | |||
==== Tyrosine Phosphorylation Mechanism ==== | ==== Tyrosine Phosphorylation Mechanism ==== | ||
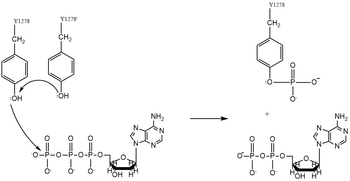
| - | Dimerization of anaplastic lymphoma kinase activates the kinase | + | [[Image:ALK_Tyrosine_Phosphorylation.png|350 px|left|thumb|Figure 4. Tyrosine phosphorylation mechanism]] |
| + | |||
| + | Dimerization of anaplastic lymphoma kinase activates the <scene name='90/904309/Kinase_domain/2'>kinase domain</scene> of each monomer <ref name="Li">PMID:34819665</ref>. Next, the kinase domains phosphorylate the [https://en.wikipedia.org/wiki/Tyrosine_phosphorylation tyrosine residues] (Y1278, Y1282, and Y1283 <ref name="Hallberg">PMID:24060861</ref>) of the opposite monomer using the hydrolysis of ATP into ADP and Pi <ref name="Munck">PMID:34646012</ref>. These phosphorylated tyrosine residues recruit signal proteins via phosphorylation mechanisms similar to that of the IR. These signal proteins start signaling cascades through various signal pathways including the ERK, JAK, and PI3K pathways. These pathways signal for cell proliferation and survival (ex: begin transcription). The mechanism of tyrosine phosphorylation is a key step in the signal activation, transduction, and regulation of ALK enzymatic activity. This mechanism is similar to Class II RTKs, used by the IR in insulin signal activation and transduction. Mutations in catalytic tyrosine residues leads to non-functional ALK <ref name="Huang">PMID: 30400214</ref>. In the absence of a bound ligand, it is proposed that the kinase domain of ALK is instead cleaved by Caspase 3 (Casp 3,) which subsequently induces apoptosis <ref name="Palmer">PMID: 19459784</ref>. | ||
| + | |||
== Applications == | == Applications == | ||
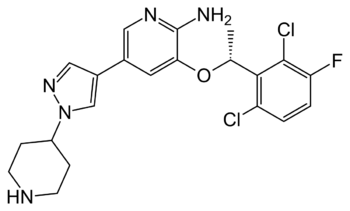
| - | + | [[Image:Crizotinib_Structure.png|350 px|right|thumb|Figure 5. Structure of the ALK inhibitor, Crizotinib]] | |
| + | Characterization of ALK as a proto-oncogene implicated in various types of cancers has made it an attractive target for drug therapies <ref name="Lewis">PMID:22734674</ref>. Treatments are designed to correct overactivation of the kinase domain of ALK, one of the main observations in ALK-positive cancers. ALK-positive cancers are those in which the individual has an oncogenic mutation in the ALK protein sequence that contributes to the proliferation of cancer cells. Overactivation of the kinase domain is caused by aberrant ATP binding, leading to the upregulation of cell growth signaling pathways. This overexpression of growth signals allows cancerous cells to grow and divide more rapidly than normal cells. One therapeutic medication seeing increased use with high levels of success is <scene name='90/904309/Crizotinib/1'>Crizotinib</scene>. Approved by the FDA in January of 2021, Crizotinib treats pediatric/young adult ALK-positive anaplastic large cell lymphoma. The treatment has an overall response rate of 90%, working by <scene name='90/904309/Crizotinib_bound/1'>binding</scene> to the ATP binding site of the kinase domain <ref name="Sahu">PMID:24455567</ref>. Crizotinib preferentially binds to ATP in this region, therefore effectively blocking the binding of ATP to the kinase domain <ref name="Sahu" />. This prevents the signal transduction via intercellular signaling pathways, which works to prevent cancerous cells from growing and spreading. Other possible inhibition mechanisms of Crizotinib suggest the inhibition of the RTK, [https://en.wikipedia.org/wiki/C-Met cMET], as the ATP binding site of both kinase domains being very similar both sequentially and structurally <ref name="Wang">PMID:25399641</ref>. Apart from possible inhibition of cMET, Crizotinib is regarded as a highly successful in regard to its specificity for the ATP binding site of the ALK kinase domain <ref name="Wang">PMID:25399641</ref>. | ||
</StructureSection> | </StructureSection> | ||
| Line 25: | Line 37: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | ==PDB Files Used== | ||
| + | [https://www.rcsb.org/structure/7N00 7N00], [https://www.rcsb.org/structure/4ANQ 4ANQ],[https://www.rcsb.org/structure/3LCS 3LCS],[https://www.rcsb.org/structure/2XP2 2XP2] | ||
== Student Contributors == | == Student Contributors == | ||
*Kaylin Todor | *Kaylin Todor | ||
*Rebekah White | *Rebekah White | ||
Current revision
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Reshetnyak AV, Rossi P, Myasnikov AG, Sowaileh M, Mohanty J, Nourse A, Miller DJ, Lax I, Schlessinger J, Kalodimos CG. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature. 2021 Dec;600(7887):153-157. doi: 10.1038/s41586-021-04140-8. Epub 2021, Nov 24. PMID:34819673 doi:http://dx.doi.org/10.1038/s41586-021-04140-8
- ↑ 2.0 2.1 2.2 2.3 Huang H. Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase: A Catalytic Receptor with Many Faces. Int J Mol Sci. 2018 Nov 2;19(11). pii: ijms19113448. doi: 10.3390/ijms19113448. PMID:30400214 doi:http://dx.doi.org/10.3390/ijms19113448
- ↑ 3.0 3.1 3.2 Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochem J. 2009 May 27;420(3):345-61. doi: 10.1042/BJ20090387. PMID:19459784 doi:http://dx.doi.org/10.1042/BJ20090387
- ↑ Li T, Stayrook SE, Tsutsui Y, Zhang J, Wang Y, Li H, Proffitt A, Krimmer SG, Ahmed M, Belliveau O, Walker IX, Mudumbi KC, Suzuki Y, Lax I, Alvarado D, Lemmon MA, Schlessinger J, Klein DE. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature. 2021 Dec;600(7887):148-152. doi: 10.1038/s41586-021-04141-7. Epub 2021, Nov 24. PMID:34819665 doi:http://dx.doi.org/10.1038/s41586-021-04141-7
- ↑ Pedersen NB, Wang S, Narimatsu Y, Yang Z, Halim A, Schjoldager KT, Madsen TD, Seidah NG, Bennett EP, Levery SB, Clausen H. Low density lipoprotein receptor class A repeats are O-glycosylated in linker regions. J Biol Chem. 2014 Jun 20;289(25):17312-24. doi: 10.1074/jbc.M113.545053. Epub, 2014 May 5. PMID:24798328 doi:http://dx.doi.org/10.1074/jbc.M113.545053
- ↑ 6.0 6.1 Selander-Sunnerhagen M, Ullner M, Persson E, Teleman O, Stenflo J, Drakenberg T. How an epidermal growth factor (EGF)-like domain binds calcium. High resolution NMR structure of the calcium form of the NH2-terminal EGF-like domain in coagulation factor X. J Biol Chem. 1992 Sep 25;267(27):19642-9. PMID:1527084
- ↑ 7.0 7.1 Li T, Stayrook SE, Tsutsui Y, Zhang J, Wang Y, Li H, Proffitt A, Krimmer SG, Ahmed M, Belliveau O, Walker IX, Mudumbi KC, Suzuki Y, Lax I, Alvarado D, Lemmon MA, Schlessinger J, Klein DE. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature. 2021 Dec;600(7887):148-152. doi: 10.1038/s41586-021-04141-7. Epub 2021, Nov 24. PMID:34819665 doi:http://dx.doi.org/10.1038/s41586-021-04141-7
- ↑ 8.0 8.1 De Munck S, Provost M, Kurikawa M, Omori I, Mukohyama J, Felix J, Bloch Y, Abdel-Wahab O, Bazan JF, Yoshimi A, Savvides SN. Structural basis of cytokine-mediated activation of ALK family receptors. Nature. 2021 Oct 13. pii: 10.1038/s41586-021-03959-5. doi:, 10.1038/s41586-021-03959-5. PMID:34646012 doi:http://dx.doi.org/10.1038/s41586-021-03959-5
- ↑ Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013 Oct;13(10):685-700. doi: 10.1038/nrc3580. PMID:24060861 doi:http://dx.doi.org/10.1038/nrc3580
- ↑ Lewis RT, Bode CM, Choquette D, Potashman M, Romero K, Stellwagen JC, Teffera Y, Moore E, Whittington DA, Chen H, Epstein LF, Emkey R, Andrews PS, Yu V, Saffran DC, Xu M, Drew AE, Merkel P, Szilvassy S, Brake RL. The discovery and optimization of a novel class of potent, selective and orally bioavailable Anaplastic Lymphoma Kinase (ALK) Inhibitors with potential utility for the treatment of cancer. J Med Chem. 2012 Jun 26. PMID:22734674 doi:10.1021/jm3005866
- ↑ 11.0 11.1 Sahu A, Prabhash K, Noronha V, Joshi A, Desai S. Crizotinib: A comprehensive review. South Asian J Cancer. 2013 Apr;2(2):91-7. doi: 10.4103/2278-330X.110506. PMID:24455567 doi:http://dx.doi.org/10.4103/2278-330X.110506
- ↑ 12.0 12.1 Wang Q, Zorn JA, Kuriyan J. A structural atlas of kinases inhibited by clinically approved drugs. Methods Enzymol. 2014;548:23-67. doi: 10.1016/B978-0-12-397918-6.00002-1. PMID:25399641 doi:http://dx.doi.org/10.1016/B978-0-12-397918-6.00002-1
PDB Files Used
Student Contributors
- Kaylin Todor
- Rebekah White