|
|
| (80 intermediate revisions not shown.) |
| Line 3: |
Line 3: |
| | | | |
| | <StructureSection load='7PGR' size='350' frame='true' | | <StructureSection load='7PGR' size='350' frame='true' |
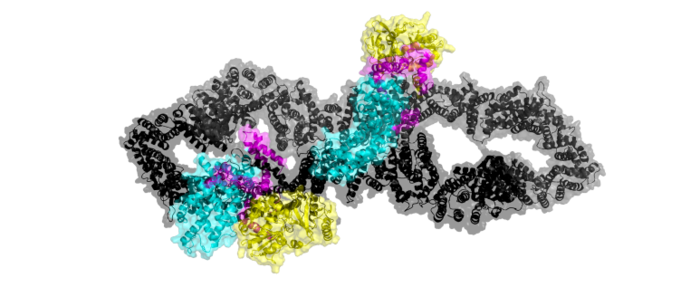
| - | side='right' caption='Neurofibromin Closed Conformation 7PGR' scene='90/904311/Open_conformation/2'> | + | side='right' caption='Structural representation of the GAP protein Neurofibromin in its open conformation. The N-C HEAT ARM of both monomers are in black. The GRD domain is in Cyan. The GAPex domain is in Magenta. The Sec14-PH domain is in Yellow. PDB code:[https://www.rcsb.org/structure/7PGT 7PGT]' scene='90/904311/Open_conformation/10'> |
| | | | |
| - | [[Image:Open_photo.png|800px|right|thumb]] | + | [[Image:Open_photo.png|700px|right|thumb]] |
| | | | |
| | ==Introduction== | | ==Introduction== |
| - | [https://en.wikipedia.org/wiki/Neurofibromatosis_type_I Neurofibromatosis Type 1] is a genetic disorder caused by mutations in the tumor suppressor gene NF1 that codes for the [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] neurofibromin. Neurofibromin is closely involved in signaling pathways such as [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MARP/ERK], [https://en.wikipedia.org/wiki/PI3K/AKT/mTOR_pathway P13K/AKT/mTOR], and other cell signaling pathways that use [https://en.wikipedia.org/wiki/Ras_GTPase Ras] <ref name="Bergoug"> DOI:10.3390/cells9112365</ref>. Mutations that cause a decrease in activity of neurofibromin cause tumors to grow along your nerves. As neurofibromin is ubiquitously expressed throughout the body, these tumors can grow anywhere. Neurofibromin is located in the [https://en.wikipedia.org/wiki/Cytosol cytosol] of the cell but is recruited to the [https://en.wikipedia.org/wiki/Cell_membrane plasma membrane] to bind to [https://en.wikipedia.org/wiki/Ras_GTPase Ras]. The structure of neurofibromin was detemed through high-resolution single particles [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryogenic electron microscopy] to understand the overall structure and the different domains. [https://en.wikipedia.org/wiki/X-ray_crystallography#:~:text=X%2Dray%20crystallography%20is%20the,diffract%20into%20many%20specific%20directions. X-Ray crystallography] experiments found inconsistency in the structural determination.<ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | + | [https://en.wikipedia.org/wiki/Neurofibromatosis_type_I Neurofibromatosis Type 1] is a genetic disorder caused by mutations in the tumor suppressor gene NF1 that codes for the [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] neurofibromin.<ref name="Bergoug"> DOI:10.3390/cells9112365</ref> Neurofibromin is closely involved in signaling pathways such as [https://en.wikipedia.org/wiki/MAPK/ERK_pathway MAPK/ERK], [https://en.wikipedia.org/wiki/PI3K/AKT/mTOR_pathway P13K/AKT/mTOR], and other cell signaling pathways that use [https://en.wikipedia.org/wiki/Ras_GTPase Ras] <ref name="Bergoug"> DOI:10.3390/cells9112365</ref>. Decreased activity of neurofibromin due to mutation can lead to tumor growth along nerves. As it is ubiquitous expression, NF1 misregulation can cause systemic tumor growth. Neurofibromin is localized to the [https://en.wikipedia.org/wiki/Cytosol cytosol] but is recruited to the [https://en.wikipedia.org/wiki/Cell_membrane plasma membrane] to inactivate [https://en.wikipedia.org/wiki/Ras_GTPase Ras]. The structure of neurofibromin was determined by high-resolution single particle [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryo-EM].<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> These structures illustrated the domain architecture and conformational changes in neurofibromin, controlling Ras binding and inactivation. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref> |
| | + | |
| | ==Function== | | ==Function== |
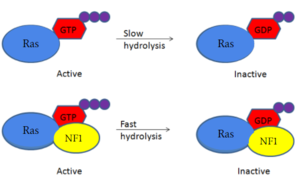
| - | Neurofibromin is a [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] that binds to [https://en.wikipedia.org/wiki/Ras_GTPase Ras], a [https://en.wikipedia.org/wiki/GTPase GTPase], to increase the hydrolysis of GTP to GDP. This inactivates the cell signaling of Ras until another [https://en.wikipedia.org/wiki/Guanosine_triphosphate GTP] can replace the [https://en.wikipedia.org/wiki/Guanosine_diphosphate GDP] from the cytosol. Neurofibromin and Ras binding is possible in only the <scene name='90/904311/Open_conformation/2'>open conformation</scene> of Neurofibromin. The mechanism is shown in figure 1 and displays the slow hydrolysis of GTP bound to Ras and the fast hydrolysis of GTP when bound to Neurofibromin. <ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> [[Image:RasNeurofibrominMech.PNG|300px|left|thumb|Figure 1: Simple diagram of Ras GTP hydrolysis when bound to NF1 and unbound. The speed of GTP hydrolysis is significantly increased when bound to NF1. Ras is inactive when bound to GDP and active when bound to GTP ]] | + | Neurofibromin is a [https://en.wikipedia.org/wiki/GTPase-activating_protein GTPase-activating protein] that binds to [https://en.wikipedia.org/wiki/Ras_GTPase Ras], a [https://en.wikipedia.org/wiki/GTPase GTPase], to increase its inherent hydrolysis of GTP to GDP (Figure 1).<ref name="Bourne"> DOI:10.1038/39470</ref> This inactivates the cell signaling of Ras until reactivated by [https://en.wikipedia.org/wiki/Guanosine_triphosphate GTP] exchange. Neurofibromin has two structural conformations, the open and closed conformation. Neurofibromin only binds to Ras in its open conformation. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> [[Image:RasNeurofibrominMech.PNG|300px|right|thumb|Figure 1: Shifted rate of of Ras GTP hydrolysis when bound to neurofibromin. The speed of GTP hydrolysis is significantly increased when bound to neurofibromin. Ras is inactive when bound to GDP and active when bound to GTP ]] |
| | + | |
| | ==Structure== | | ==Structure== |
| - | Neurofibromin is a [https://en.wikipedia.org/wiki/Protein_dimer protein dimer] that exists in the<scene name='90/904311/Closed_conformation/3'>closed</scene> and <scene name='90/904311/Open_conformation/2'>open</scene> conformation. Each [https://en.wikipedia.org/wiki/Protomer protomer] contains a GRD, Sec14-PH, and a GAPex domain located on a HEAT N-C arm. Ras binds to the GRD site with Arg1276 being the critical residue for binding. <ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | + | Neurofibromin is a [https://en.wikipedia.org/wiki/Protein_dimer protein dimer] that exists in <scene name='90/904311/Closed_conformation/8'>closed</scene> and <scene name='90/904311/Open_conformation/14'> open</scene> conformations. The exact initiation of structural rearrangement is currently unknown and a point of further research. <ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> <ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>. Both protomers contain 5 separate domains, a N-HEAT/ARM, C-HEAT/ARM, GRD binding site, GAPex domain, and Sec14-PH domain. |
| - | ===Closed conformation=== | + | |
| - | Ras is unable to bind to the GRD active site when both of the NF1 protomers are in the closed confirmation. In the<scene name='90/904311/Closed_conformation/3'>closed conformation</scene>, one protomer has its domains shifted by a 130 degree rotation of three separate linkers. That rotation places <scene name='90/904311/Closed_arg/4'>Arg1276 in the closed conformation</scene> in an orientation that <scene name='90/904312/Closed_zoom/1'>sterically hinders the binding between Ras and Arg1276</scene> in the GRD site (Figure 3). Also in this depiction Ras binding to the GRD site is inhibited by the N-HEAT ARM as it interferes at the GRD in Ras attempting to bind to the active site and is sterically hindered. Making <scene name='90/904311/Closed_with_ras/2'>Ras binding in closed conformation</scene> sterically impossible in the <scene name='90/904311/Closed_conformation/3'>closed conformation</scene>
| + | ===Domains=== |
| - | . The <scene name='90/904311/Closed_conformation/3'>closed conformation</scene> can exist naturally without any form of stabilization but will also fall back to the <scene name='90/904311/Open_conformation/2'>open conformation</scene>.<ref name="Bourne"> DOI:10.1038/39470</ref> <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
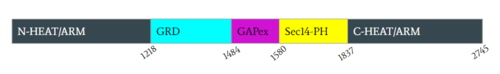
| + | [[Image:Domain.PNG|500px|none|thumb|Figure 2: Domains for NF1 with their location on the <scene name='90/904311/Domainintro_open/11'>neurofibromin monomer</scene> (open conformation) and the pertinent colors. Numerical values show domain locations along the polypeptide chain.]] |
| - | ====Zinc Stabilized==== | + | |
| - | The <scene name='90/904311/Closed_conformation/3'>closed conformation</scene> of Neurofibromin can be stabilized by a zinc ion to prevent the shift back to an <scene name='90/904311/Open_conformation/2'>open conformation</scene>. This binding is done between <scene name='90/904311/Zinc_binding_site/5'>Cys1032, His1558, His1576, and Zinc</scene>. Cys1032 is within the N-HEAT domain and both His1558 and His1576 are within the GAPex-subdomain. When zinc stabilizes Neurofibromin, it will stay in the <scene name='90/904311/Closed_conformation/3'>closed conformation</scene> and continue to inhibit the binding of RAS.<ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> | + | ====N-C HEAT/ARM==== |
| - | ===Open conformation=== | + | The N-C HEAT/ARM are two separate but similar connected domains that make up the backbone of the protomer; <scene name='90/904311/N-heatarm_open/2'>N-HEAT/ARM</scene>(N-Terminal) and <scene name='90/904311/C-heatarm_open/2'>C-HEAT/ARM</scene>(C-Terminal). [https://en.wikipedia.org/wiki/HEAT_repeat HEAT] and [https://en.wikipedia.org/wiki/Armadillo_repeat ARM] are acronyms for a trope of repeating helical structures. Structurally, the N-HEAT/ARM consists of various helices and loops that interconnect on one of the existing protomers. The N-HEAT/ARM is critical in stabilization and linking the catalytic and core domains.<ref name="Bergoug"> DOI:10.3390/cells9112365</ref> The N-HEAT/ARM in its linkage with the GRD catalytic domain also has a direct impact on the relative conformational change from the active (Ras BOUND) to inactive (Ras unbound) states. The Zn2+ binding site also extends from the N-HEAT ARM just before the GRD and SEC14-PH domains.<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> The C-HEAT/ARM plays similar roles to the N-HEAT/ARM by stabilizing and linking the GRD and SEC14-PH domain and contributing to the conformational changes from the closed to open state.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref> |
| - | In the open state, one of the protomers remains in the closed confirmation inaccessible to Ras, while the other protomer is conformationally changed into the open form with Ras bound.
| + | |
| - | In the <scene name='90/904311/Open_conformation/2'>open conformation</scene> the one protomer is shifted due to a 90 rotation. This rotation allows for binding between RAS and the <scene name='90/904311/Arg_1276_open/1'>Arg1276</scene>
| + | ====GRD==== |
| - | in the GRD site while in the <scene name='90/904311/Open_conformation/2'>open conformation</scene>. Allowing for the <scene name='90/904312/Arg_1276_open/11'>Arg1276 interaction with Ras in the open conformation</scene> to occur without any steric hindrance as shown in the <scene name='90/904311/Closed_conformation/3'>closed conformation</scene>. In the open state, one of the protomers remains in the closed confirmation inaccessible to Ras, while the other protomer is conformationally changed into the open form with Ras bound. It is important to mention, the GRD and Sec14-PH are reoriented away from one another and the GRD site is no longer sterically hindered and clashing with the N-HEAT ARM and is completely accessible for Ras to bind. To undergo the movement to the open confirmation, significant conformational changes exist within three separate linkers (L1, L2, L3).<ref name="Bourne"> DOI:10.1038/39470</ref> <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | Neurofibromin’s main catalytic domain is the <scene name='90/904311/Grd_open/2'>GRD</scene> binding site. Linked structurally to both HEAT/ARMs, it consists of mainly loops and helices. There is one single GRD binding site per protomer. In the closed state, Ras cannot bind the GRD site due to a steric hindrance in which Ras clashes with the N-HEAT/ARM.<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
| - | ===Conformational Change Linkers===
| + | Within the GRD site, the critical binding site residue is Arg1276. GRD also can bind to SPRED-1 to be recruited from the cytosol to the plasma membrane. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref> |
| - | The rotation of the domains between the <scene name='90/904311/Open_conformation/2'>open conformation</scene> and <scene name='90/904311/Closed_conformation/3'>closed conformation</scene> of Neurofibromin are conducted by three helical linkers named L1, L2, and L3. The <scene name='90/904311/Linker_closed/1'>linkers in the closed conformation</scene>
| + | ====GAPex==== |
| - | <scene name='90/904312/Linker_closed/2'>(zoomed in)'</scene> and the <scene name='90/904311/Linkers_open/1'>linkers in the open conformation</scene> | + | The <scene name='90/904311/Gapex_open/2'>GAPex</scene> of the GRD site lies between the Sec14-PH and GRD catalytic sites. This domain is non-catalytic and structurally consists of various loops and helices. Its may contribute to the binding of SPRED-1 to the GRD site. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
| - | <scene name='90/904312/Zoomed_lo/1'>(zoomed in)'</scene> undergo rotations to relocate the GRD and Sec14-PH sites. Linker 1 (L1) consists of a loop connected by two helices from L1173-M1215 and is the main contributor in rotation of the GRD domain. The rotation of L1 to the <scene name='90/904311/Linkers_open/1'>open conformation</scene> causes N-HEAT ARM [https://en.wikipedia.org/wiki/Alpha_helix alpha helix] 48 and GRD helix 49 to extend out, aligning to form a hinge point at G1190. The GRD relocation is assisted by Sec14-PH relocation, which is initiated by Linker 3(L3) from Q1835 to G1852 where the proline rich section of the C-HEAT ARM changes conformation.L1 and L3 move closer to each other in the <scene name='90/904311/Linkers_open/1'>open conformation</scene> allowing for the GRD and Sec14-PH domain to initiate their significant rearrangement. Linker 2 (L2) consists of residues G1547-T1565 and begins at helix 63, the final helix of the GRD site, and connects into the short loop of [https://en.wikipedia.org/wiki/Alpha_helix alpha helix] 65 of the Sec14-PH domain and also assists in shifting the Sec14-PH away from the GRD site. The combination of these three linkers are responsible for the conformational shift of the <scene name='90/904311/Linker_closed/1'> closed conformation</scene> | + | ====Sec14-PH==== |
| - | <scene name='90/904312/Linker_closed/2'>(zoomed in)'</scene> and <scene name='90/904311/Linkers_open/1'>open conformation</scene>
| + | The <scene name='90/904311/Sec14ph_open/2'>Sec14-PH </scene> is linked and extends out from the HEAT/ARM’s. Sec14-PH functions as a membrane associated domain and using a hydrophobic cavity binds to neurofibromin the plasma membrane. The closed conformation also blocks Sec14-PH binding to the cell membrane due to blocking by GRD. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
| - | <scene name='90/904312/Zoomed_lo/1'>(zoomed in)'</scene>.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | |
| - | ==Domains==
| + | ===Conformations=== |
| - | [[Image:Domain.PNG|500px|right|thumb|Figure 2: Domains for NF1 and the pertinent colors. Numerical values show domain locations along the polypeptide chain.]]
| + | |
| - | ===N-C HEAT ARM===
| + | ====Closed conformation==== |
| - | NF1 in its closed and open confirmations holds five separate domains. The first domain shown in black is representative of two separate but very similar domains, the N-HEAT ARM(N-Terminal) and C-HEAT ARM(C-Terminal). Structurally the N-HEAT Arm’s consists of various helices and loops that interconnect on one of the existing protomers. The N-HEAT ARM is critical in stabilization and linking the other domains which extend out from the core and are important to its catalytic function. The N-HEAT ARM in its linkage with the GRD catalytic domain, has a direct impact on the relative conformational change from the active (Ras BOUND) to inactive (Ras unbound) states. Finally, the Zn2+ binding site also exists downstream on the N-HEAT ARM just before the GRD and SEC14-PH domains. The C-HEAT Arm is linked structurally to the N-HEAT ARM and is a continuation of the protomer. It extends out on the opposing end from the other domains. In its structure consisting of various loops and helices, it is involved with stabilization and linking to the GRD and SEC14-PH and plays a role in the conformational changes from the closed to open state.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | Ras is unable to bind to the GRD binding site when both of the neurofibromin protomers are in the <scene name='90/904311/Closed_conformation/8'>closed conformation</scene>. In the closed conformation, one protomer has its domains shifted by a 130° rotation of three separate conformational change linkers.<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> That rotation places Arg1276 in an orientation where binding of Y32 of Ras is sterically hindered by E31.<ref name="Bourne"> DOI:10.1038/39470</ref> (<scene name='90/904311/Closed_arg/6'>Arg1276, Y32, and E31 interaction</scene> |
| - | ===GRD=== | + | <jmol> |
| - | Neurofibromin’s main catalytic domain is the GRD active site. Linked structurally to both HEAT ARM’s, it consists of mainly loops and helices. Per protomer, there is one single GRD binding site. In the closed state Ras cannot bind due to a steric hindrance in which Ras clashes with the N-HEAT ARM upon attempting to bind to the GRD site. In its active state GRD can bind Ras. The critical residue within the GRD site is Arg1276.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | <jmolButton> |
| - | ===GAPex-Subdomain===
| + | <script>moveto 1.0 { 347 936 -57 112.47} 2508.34 0.0 0.0 {366.9273333333333 357.0333333333333 434.887} 189.81815871562094 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> |
| - | The GAPex subdomain of the GRD site lies between the Sec14-PH and GRD catalytic sites. This domain is non-catalytic and structurally consists of various loops and helices. Its main function is to bind SPRED-1, which is a recruiter protein that binds to this subdomain in the cytosol to recruit Neurofibromin to the plasma membrane. <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | </jmolButton> |
| - | ===Sec14-PH===
| + | </jmol>) Ras binding to the GRD site is inhibited by steric occlusion from the N-HEAT/ARM. The closed conformation can exist naturally without any form of stabilization but exists in a natural equilibrium with the open conformation. |
| - | The Sec14-PH domain is linked and extends out from the HEAT ARM’s and consists of largely various helices and loops. Its function is a membrane associated domain and holds a largely hydrophobic cavity allowing for binding to the plasma membrane. In the <scene name='90/904311/Closed_conformation/3'>closed conformation</scene> it is blocked by the GRD and is inaccessible to the lipid membrane. In the <scene name='90/904311/Open_conformation/2'>open conformation</scene> it becomes exposed and can access the lipid membrane for interaction.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | |
| - | ==Arginine 1276==
| + | =====Zinc Stabilized===== |
| - | Arg1276 is the critical residue within the GRD site needed for proper binding to Ras. The interaction between <scene name='90/904311/Arg_1276_open/1'>Arg1276 and Ras in the open conformation</scene>
| + | The closed conformation is stabilized by a zinc ion that prevents the shift back to the open conformation. Zinc binding is chelated by three residues (<scene name='90/904311/Zinc_binding_site/11'>Cys1032, His1558, His1576</scene>). These three residues are contributed by the N-HEAT domain (Cys1032) and the GAPex-subdomain (His1558 and His1576). Zinc stabilization keeps Neurofibromin in the closed conformation, inhibiting Ras binding.<ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
| - | <scene name='90/904312/Arg_1276_open/11'>(zoomed in)</scene> is only possible in the open confirmation (Figure 4). In the open confirmation, the arginine finger binds to the backbone γ-carbon of Y32 to assist in eventual hydrolysis of GTP. In the closed conformation this was inhibited by E31 but in the open conformation the conformational change allows for an interaction of R1276 and the γ-carbon so they align with one another. This allows for the normal function of NF1 in the rapid rate increase of GTP hydrolysis upon Ras binding. | + | |
| - | <scene name='90/904311/Closed_arg/4'>Arg1276 in closed conformation </scene> | + | ====Open conformation==== |
| - | <scene name='90/904312/Closed_zoom/1'>(zoomed in)</scene> has steric clashing with Ras making binding of Ras impossible (Figure 3). In the closed confirmation the Arginine finger attempts to bind the backbone gamma carbon(RAS) of a Tyrosine residue(Y32) to assist in hydrolysis, but instead clashes with a Glutamate residue(E31) that would otherwise not interfere in the open conformation and can no longer bind properly. This residue impacts binding of Ras and is of critical importance in determining if Ras is able to bind to the GRD site or not to assist in eventual hydrolysis of GTP. | + | In the <scene name='90/904311/Open_conformation/14'> open conformation</scene>, one protomer is shifted to allow Ras binding, while the other protomer remains in the closed conformation. In the open conformation one protomer is rotated 90°, facilitating binding between Y32 of RAS and Arg1276.<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref><ref name="Bourne"> DOI:10.1038/39470</ref> (<scene name='90/904311/Arg_1276_mi/2'>Y32 and Arg1276 interaction</scene> |
| - | Mutations to the arginine finger are shown to slow function of NF1 and other GTPases in their function in accelerating GTP hydrolysis. When a Ras GAP such as NF1 can functionally utilize its arginine finger, it allows NF1 to rapidly hydrolyze GTP when bound to Ras. This emphasizes the importance of the R1276 residue for the GRD site binding to Ras.<ref name="Bourne"> DOI:10.1038/39470</ref><ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | <jmol> |
| - | [[Image:ClosedArgFinger.PNG|400px|left|thumb| Figure 3: Arg1276 of the GRD active site is sterically hindered in the closed conformation by Glu31 of Ras as it attempts to interact with the γ-carbon of Tyr32, but is unable to properly form a bond. Making binding to Ras impossible in the closed conformation. ]] [[Image:OpenArgFinger.PNG|400px|none|thumb|Figure 4: Arg1276 of the GRD active site is no longer sterically hindered in the open conformation by Glu31 of Ras, making it able to interact with γ-carbon of Tyr32 and form a bond with Ras. ]]
| + | <jmolButton> |
| | + | <script>moveto 1.0 { 294 -950 -102 45.37} 2089.03 0.0 0.0 {360.8673666666667 403.1638666666667 362.0271666666667} 189.97419116715565 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> |
| | + | </jmolButton> |
| | + | </jmol>) |
| | + | in the GRD site. To allow Ras binding, the GRD and Sec14-PH domains are reoriented away from one another and the GRD site is accessible for Ras binding. This conformational change to the open conformation is driven by rearrangement of three separate linkers (L1, L2, L3). |
| | + | |
| | + | ====Conformational Change Linkers==== |
| | + | These three helical linkers (L1, L2, and L3) undergo rotations to relocate the GRD and Sec14-PH sites (<scene name='90/904311/Linkers_open/2'>linkers in open conformation</scene> |
| | + | <jmol> |
| | + | <jmolButton> |
| | + | <script>moveto 1.0 { 258 942 215 149.04} 254.03 0.0 0.0 {363.6198596926281 345.1996488441136 383.09000113650916} 143.7868028412415 {0 0 0} 0 0 0 3.0 0.0 0.0 </script> <text>🔎</text> |
| | + | </jmolButton> |
| | + | </jmol> and <scene name='90/904311/Linker_closed/4'>linkers in closed conformation</scene> |
| | + | <jmol> |
| | + | <jmolButton> |
| | + | <script>moveto 1.0 { -93 -976 -198 100.01} 317.4 0.0 0.0 {364.4077734377054 341.9358671885079 392.3046403678749} 146.92471247967674 {0 0 0} 0 0 0 3.0 0.0 0.0 </script> <text>🔎</text> |
| | + | </jmolButton> |
| | + | </jmol>). Linker 1 (L1) consists of a loop connected by two helices from L1173-M1215 and is the main contributor in rotation of the GRD domain. The rotation of L1 to the open conformation causes N-HEAT/ARM [https://en.wikipedia.org/wiki/Alpha_helix α helix] 48 and GRD helix 49 to extend out, aligning to form a hinge point at G1190.<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
| | + | The GRD relocation is assisted by Sec14-PH relocation, which is initiated by Linker 3(L3) from Q1835 to G1852. Movement of L3 is further supported by rearrangement of the proline rich section of the C-HEAT/ARM. L1 and L3 also move closer to each other in the open conformation initiating rearrangement of the GRD and Sec14-PH domain. Linker 2 (L2) consists of residues G1547-T1565 and begins at helix 63, the final helix of the GRD site, and connects into the short loop of [https://en.wikipedia.org/wiki/Alpha_helix α helix] 65 of the Sec14-PH domain and also assists in shifting the Sec14-PH away from the GRD site.<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> The combination of these three linkers are largely responsible for the conformational shift of the closed and open conformation.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref> |
| | + | ==Ras Binding== |
| | + | Arg1276 is the critical residue within the GRD site for Ras activation. When <scene name='90/904311/Arg_1276_open/13'>Arg1276 is in the open conformation</scene> |
| | + | <jmol> |
| | + | <jmolButton> |
| | + | <script>moveto 1.0 { -776 433 -458 146.98} 7209.11 0.0 0.0 {361.5375833333334 402.0207083333333 360.750125} 190.41144084626742 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> |
| | + | </jmolButton> |
| | + | </jmol>, the arginine finger binds to the backbone γ-carbon of Y32 in Ras to assist in eventual hydrolysis of GTP.<ref name="Bourne"> DOI:10.1038/39470</ref> When <scene name='90/904311/Closed_zoom/10'>Arg1276 is in the closed conformation</scene> |
| | + | <jmol> |
| | + | <jmolButton> |
| | + | <script>moveto 1.0 { -816 -263 -515 73.84} 7673.3 0.0 0.0 {369.48317647058826 352.12982352941174 432.1719411764706} 186.8074765770222 {0 0 0} 0 0 0 3.0 0.0 0.0</script> <text>🔎</text> |
| | + | </jmolButton> |
| | + | </jmol> the interaction between Y32 and GRD (R1276) is blocked by E31. Removal of the inhibition of E31 from Ras by R1276 from Neurofibromin allows for the normal function of neurofibromin in the rapid rate increase of GTP hydrolysis upon Ras binding.<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> Mutations to the arginine finger slow GTPase activating reaction of neurofibromin.<ref name="Bourne"> DOI:10.1038/39470</ref> |
| | ==SPRED 1== | | ==SPRED 1== |
| - | SPRED 1 is a protein that binds to the GAPex domain of Neurofibromin. Its function recruits the Neurofibromin protein when bound from the cytosol to the plasma membrane. SPRED 1 will bind to the GAPex domain of Neurofibromin in the <scene name='90/904311/Sped-1_closed/1'>closed conformation'</scene> in the cytosol to recruit Neurofibromin to the plasma membrane. Unlike Ras, SPRED-1 does show the ability to <scene name='90/904311/Spred-1_open/1'>bind to the open conformation</scene>. When bound to the open conformation of Neurofibromin in the cytosol, it may present a different orientation that impacts the recruitment to the plasma membrane. Further research is needed to assess the impact of the function and the changes it may present.<ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref><ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | <scene name='90/904311/Spred1_w_grd/2'>SPRED 1</scene> is another peripheral protein that interacts with the GRD domain of Neurofibromin. SPRED 1 recruits Neurofibromin from the cytosol to the plasma membrane to interact with Ras. Binding of SPRED 1 can occur in either the open or closed conformation and causes a structural rearrangement of the GRD and GAPex domains that has yet to be structuralized.<ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref> |
| - | ==Future Clinical Relevance== | + | |
| - | [https://en.wikipedia.org/wiki/Germline_mutation Germline mutations] are common in NF1, often causing genetic tumor syndrome through misregulation of the Ras signaling pathway. [https://en.wikipedia.org/wiki/Somatic_mutation Somatic mutations] among NF1 are also extremely common. In germline mutations and some somatic mutations of NF1, clinical findings show tumors that develop often along the deep epidermis layer of the skin. The most common among NF1 mutations is [https://en.wikipedia.org/wiki/Neurofibroma Neurofibroma]. Physically, soft skin-colored pink papules develop on the extremities or on the neck. Also in patients they become prevalent during puberty. Among germline mutations Neurofibroma seems to be the most well known, other tumors may also be common. [https://en.wikipedia.org/wiki/Lisch_nodule Lisch Nodules] are also common in the development of [https://en.wikipedia.org/wiki/Melanoma melanoma] within melanocytes as a result of NF1 mutations. Plexiform Neurofibroma and [https://en.wikipedia.org/wiki/Glaucoma Optic Glioma] also are tumor based syndromes resulting from these germline mutations. Increased risk in cancer for patients with mutations to NF1 is about 4 fold greater. Treatment options do exist for patients involving a multitude of drug based treatments coupled with chemotherapy. Surgery and laser treatments are also common. | + | ==Clinical Relevance== |
| - | Mutations to NF1 in the critical interaction involved in the GRD active site with R1276, structurally and functionally prove why they can cause cancer, as Ras is no longer mediated. The importance of this residue when binding to Ras may be able to contribute to future studies in potentially mimicking this arginine finger when bound to Ras, in cases where mutations would impact R1276 in its normal function. Observing the nature of NF1 can further drug development and further improve what is already known about NF1.<ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref><ref name="Sabatini"> DOI:10.1007/s11940-015-0355-4</ref>
| + | [https://en.wikipedia.org/wiki/Germline_mutation Germline mutations] are common in NF1 and often cause genetic tumor syndrome through misregulation of the Ras signaling pathway. <ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref> [https://en.wikipedia.org/wiki/Somatic_mutation Somatic mutations] among NF1 are also extremely common. In germline mutations and some somatic mutations of NF1, tumors develop along the deep epidermis layer of the skin. Understanding how mutations affect the structure and function of neurofibromin will allow for advancements in treatment. |
| - | ==References placeholders==
| + | <ref name="Bergoug"> DOI:10.3390/cells9112365</ref><ref name="Sabatini"> DOI:10.1007/s11940-015-0355-4</ref> |
| - | This section will be removed it is just for copy and paste purposes. We will add the PDB code names as references in the captions of each scene.
| + | |
| - | <ref name="Bergoug"> DOI:10.3390/cells9112365</ref> | + | |
| - | <ref name="Bourne"> DOI:10.1038/39470</ref>
| + | |
| - | <ref name="Kioro"> DOI:10.1038/labinvest.2016.142</ref>
| + | |
| - | <ref name="Lupton"> DOI:10.1038/s41594-021-00687-2</ref>
| + | |
| - | <ref name="Naschberger"> DOI:10.1038/s41586-021-04024-x</ref>
| + | |
| - | <ref name="Sabatini"> DOI:10.1007/s11940-015-0355-4</ref> | + | |
| | ==References== | | ==References== |
| | <references/> | | <references/> |