Sandbox Reserved 1715
From Proteopedia
(Difference between revisions)
| (13 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
== Introduction == | == Introduction == | ||
| - | Within the [https://en.wikipedia.org/wiki/Central_nervous_system central nervous system (CNS)], various [https://en.wikipedia.org/wiki/Cell_surface_receptor membrane receptors] exist to detect extracellular signaling molecules and communicate this information intracellularly. Found in [https://en.wikipedia.org/wiki/Eukaryote eukaryotes] and known for its seven transmembrane helices, [https://en.wikipedia.org/wiki/G_protein-coupled_receptor G-protein coupled | + | Within the [https://en.wikipedia.org/wiki/Central_nervous_system central nervous system (CNS)], various [https://en.wikipedia.org/wiki/Cell_surface_receptor membrane receptors] exist to detect extracellular signaling molecules and communicate this information intracellularly. Found in [https://en.wikipedia.org/wiki/Eukaryote eukaryotes] and known for its seven transmembrane helices, [https://en.wikipedia.org/wiki/G_protein-coupled_receptor G-protein coupled receptors] (GPCRs) are one type of membrane bound receptors with conserved intracellular signaling via a heterotrimeric [https://en.wikipedia.org/wiki/G_protein#:~:text=G%20proteins%2C%20also%20known%20as,a%20cell%20to%20its%20interior. G-protein]<ref name="Katritch">PMID:23140243</ref>. The metabotropic glutamate receptor (mGlu), a Class C GPCR, is a receptor utilized in glutamate signaling– which is essential in synaptic plasticity as well as the development and repair of the CNS<ref name="Niswender">PMID:20055706</ref>. These receptors are specifically found in the pre- and postsynaptic [https://en.wikipedia.org/wiki/Neuron#:~:text=A%20neuron%20or%20nerve%20cell,animals%20except%20sponges%20and%20placozoa. neurons] of the CNS<ref name="Niswender">PMID:20055706</ref>. Eight different mGlu subtypes exist which are divided into three groups (I, II, III)<ref name="Niswender">PMID:20055706</ref>. While each mGlu has a slightly different function and location, the structures of the different mGlu subtypes are very similar (Table 1) <ref name="Niswender">PMID:20055706</ref>. For each group, binding of the [https://en.wikipedia.org/wiki/Glutamate_(neurotransmitter) neurotransmitter glutamate] to the mGlu introduces a conformational change that can activate a G-protein<ref name="Niswender">PMID:20055706</ref>. |
{| class="wikitable" style="text-align:center;" | {| class="wikitable" style="text-align:center;" | ||
| Line 31: | Line 31: | ||
|} | |} | ||
| - | [[Image:Glutamate.png|300 px|right|thumb|Figure 1. Structure of glutamate under physiological conditions (pH 7.4).]] To activate the mGlu transformation, glutamate acts as the protein's main [https://en.wikipedia.org/wiki/Agonist agonist]. [https://en.wikipedia.org/wiki/Glutamic_acid Glutamate] is an acidic, polar [https://en.wikipedia.org/wiki/Amino_acid amino acid] (Figure 1). This agonist binds to the extracellular portion of the glutamate receptor causing the transmembrane spanning region of the homodimer to change conformationally. This change allows for the mGlu to bind to a G-protein. This binding allows for the activation of a G protein which initiates a signaling cascade within the cell that can ultimately lead to a difference in the synapse’s excitability<ref name="Seven">PMID:34194039</ref>. In mGlu, the binding affinity of glutamate is also controlled by the binding of either a positive (PAM) or negative (NAM) [https://en.wikipedia.org/wiki/Allosteric_modulator allosteric modulator] to a [https://en.wikipedia.org/wiki/Binding_site binding site] within the seven TMD<ref name="Lin">PMID:34135510</ref>. | + | [[Image:Glutamate.png|300 px|right|thumb|Figure 1. Structure of glutamate under physiological conditions (pH 7.4).]] To activate the mGlu transformation, glutamate acts as the protein's main [https://en.wikipedia.org/wiki/Agonist agonist]. [https://en.wikipedia.org/wiki/Glutamic_acid Glutamate] is an acidic, polar [https://en.wikipedia.org/wiki/Amino_acid amino acid] (Figure 1). This agonist binds to the extracellular portion of the glutamate receptor causing the transmembrane spanning region of the homodimer to change conformationally. This change allows for the mGlu to bind to a G-protein. This binding allows for the activation of a G-protein which initiates a signaling cascade within the cell that can ultimately lead to a difference in the synapse’s excitability<ref name="Seven">PMID:34194039</ref>. In mGlu, the binding affinity of glutamate is also controlled by the binding of either a positive (PAM) or negative (NAM) [https://en.wikipedia.org/wiki/Allosteric_modulator allosteric modulator] to a [https://en.wikipedia.org/wiki/Binding_site binding site] within the seven TMD<ref name="Lin">PMID:34135510</ref>. |
== Structural Highlights == | == Structural Highlights == | ||
mGlu receptors are [https://en.wikipedia.org/wiki/Protein_dimer dimeric proteins] consisting of an <scene name='90/904320/Inactive_chains/3'>alpha and beta chain</scene><ref name="Seven">PMID:34194039</ref>. While a heterodimer of different mGlu subtypes can form, only homodimeric receptors can become active<ref name="Seven">PMID:34194039</ref>. Both the alpha and beta chains are comprised of <scene name='90/904320/Mglu2_domains/9'>three domains</scene>: the venus fly trap (VFT), cysteine rich domain (CRD), and the transmembrane domain (TMD)<ref name="Seven">PMID:34194039</ref>. | mGlu receptors are [https://en.wikipedia.org/wiki/Protein_dimer dimeric proteins] consisting of an <scene name='90/904320/Inactive_chains/3'>alpha and beta chain</scene><ref name="Seven">PMID:34194039</ref>. While a heterodimer of different mGlu subtypes can form, only homodimeric receptors can become active<ref name="Seven">PMID:34194039</ref>. Both the alpha and beta chains are comprised of <scene name='90/904320/Mglu2_domains/9'>three domains</scene>: the venus fly trap (VFT), cysteine rich domain (CRD), and the transmembrane domain (TMD)<ref name="Seven">PMID:34194039</ref>. | ||
| Line 37: | Line 37: | ||
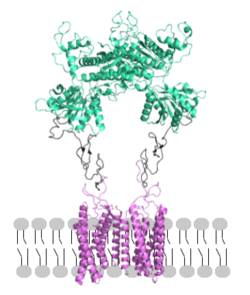
[[Image: Orientation_2.png|250px|left|thumb|Figure 2. Proper orientation of mGlu about the cell membrane.]] | [[Image: Orientation_2.png|250px|left|thumb|Figure 2. Proper orientation of mGlu about the cell membrane.]] | ||
==== Domains ==== | ==== Domains ==== | ||
| - | <scene name='90/904320/Mglu2_domains_vft/4'>VFT</scene>: The extracellular location in which the two glutamate agonists bind is known as the VFT. This domain includes a [https://en.wikipedia.org/wiki/Disulfide disulfide] bond between C121 of the alpha and beta chains. This <scene name='90/904320/Inactive_mglu/ | + | <scene name='90/904320/Mglu2_domains_vft/4'>VFT</scene>: The extracellular location in which the two glutamate agonists bind is known as the VFT. This domain includes a [https://en.wikipedia.org/wiki/Disulfide disulfide] bond between C121 of the alpha and beta chains <ref name="Du">PMID:34135509</ref>. This <scene name='90/904320/Inactive_mglu/14'>disulfide bond</scene> is shifted down in the open conformation and undergoes a <scene name='90/904320/Cys_active/3'>disulfide bond upward movement</scene> upon glutamate binding which stabilizes the <scene name='90/904319/Vft/2'>active site</scene><ref name="Du">PMID:34135509</ref>. Glutamate binds within the <scene name='90/904320/Active_site_interactions/4'>binding pocket</scene> via [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding], specifically hydrogen bonding, with R57, S143, S145, T168, and K377 of the VFT<ref name="Seven">PMID:34194039</ref>. This binding initiates a closed VFT conformation. |
| - | <scene name='90/904320/Mglu2_domains_crd/7'>CRD</scene>: The portion of the protomer that connects the VFT with the TMD is known as the CRD. Many <scene name='90/904320/Crd_cysteine/3'>disulfide bonds</scene> are located in this region between cysteines. As the connecting segment of the protein, it is critical in transmitting the conformational change caused by the binding of glutamate in the VFT to the TMD<ref name="Seven">PMID:34194039</ref>. The change resulting from the binding of glutamate brings the cysteine-rich domains of the alpha and beta chain together to alter the configuration of the seven TMD helices through its interaction with the VFT extracellular loop 2 (ECL2) <ref name="Seven">PMID:34194039</ref>. This <scene name='90/904320/Active_helices/13'>ECL2 conformational change</scene> is mediated through interactions with amino acids at the apex of the CRD (e.g. I674, P676, and P753) <ref name="Seven">PMID:34194039</ref>. | + | <scene name='90/904320/Mglu2_domains_crd/7'>CRD</scene>: The portion of the [https://en.wikipedia.org/wiki/Protomer protomer] that connects the VFT with the TMD is known as the CRD. Many <scene name='90/904320/Crd_cysteine/3'>disulfide bonds</scene> are located in this region between [https://en.wikipedia.org/wiki/Cysteine cysteines]. As the connecting segment of the protein, it is critical in transmitting the conformational change caused by the binding of glutamate in the VFT to the TMD<ref name="Seven">PMID:34194039</ref>. The change resulting from the binding of glutamate brings the cysteine-rich domains of the alpha and beta chain together to alter the configuration of the seven TMD helices through its interaction with the VFT extracellular loop 2 (ECL2) <ref name="Seven">PMID:34194039</ref>. This <scene name='90/904320/Active_helices/13'>ECL2 conformational change</scene> is mediated through interactions with amino acids at the apex of the CRD (e.g. I674, P676, and P753) <ref name="Seven">PMID:34194039</ref>. |
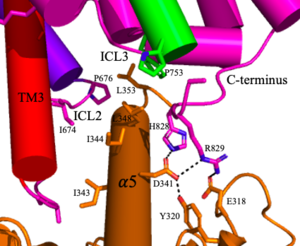
| - | <scene name='90/904320/Mglu2_domains_tmd/5'>TMD</scene>: The TMD consists of <scene name='90/904319/Inactive_tmd/9'>seven transmembrane helices</scene> that are responsible for G-protein interactions and are able to transmit the signal from ligand binding across a membrane<ref name="Niswender">PMID:20055706</ref>. In the <scene name='90/904320/Inactive_mglu2_first_picture/5'>inactive form</scene>, the asymmetric conformation of the helices is mediated by the hydrophobicity of helix 3 and 4 <ref name="Seven">PMID:34194039</ref>. This allows for a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> to form between the monomers (Figure 3A). Along with the interaction of the CRD with the ECL2 of the TMD, an allosteric modulator must bind within the transmembrane helices to allow for the conformation of the helices to be altered<ref name="Seven">PMID:34194039</ref>. This conformation allows for an <scene name='90/904320/Active_helices/14'>active dimer interface</scene> along helix 6 of both protomers (Figure 3B)<ref name="Lin">PMID:34135510</ref>. The stabilization of this conformation also enables G protein coupling with ICL2, ICL3, TM Helix 3 and the C terminus <ref name="Lin">PMID:34135510</ref> (Figure 5). | + | <scene name='90/904320/Mglu2_domains_tmd/5'>TMD</scene>: The TMD consists of <scene name='90/904319/Inactive_tmd/9'>seven transmembrane helices</scene> that are responsible for G-protein interactions and are able to transmit the signal from ligand binding across a membrane<ref name="Niswender">PMID:20055706</ref>. In the <scene name='90/904320/Inactive_mglu2_first_picture/5'>inactive form</scene>, the asymmetric conformation of the helices is mediated by the [https://en.wikipedia.org/wiki/Hydrophobe hydrophobicity] of helix 3 and 4 <ref name="Seven">PMID:34194039</ref>. This allows for a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> to form between the monomers (Figure 3A). Along with the interaction of the CRD with the ECL2 of the TMD, an allosteric modulator must bind within the transmembrane helices to allow for the conformation of the helices to be altered<ref name="Seven">PMID:34194039</ref>. This conformation allows for an <scene name='90/904320/Active_helices/14'>active dimer interface</scene> along helix 6 of both protomers (Figure 3B)<ref name="Lin">PMID:34135510</ref>. The stabilization of this conformation also enables G-protein coupling with ICL2, ICL3, TM Helix 3 and the C terminus <ref name="Lin">PMID:34135510</ref> (Figure 5). |
[[Image:Screen Shot 2022-04-19 at 2.52.21 AM.png|500px|center|thumb|Figure 3. A) The inactive transmembrane helices conformation. B) The active transmembrane helices conformation.]] | [[Image:Screen Shot 2022-04-19 at 2.52.21 AM.png|500px|center|thumb|Figure 3. A) The inactive transmembrane helices conformation. B) The active transmembrane helices conformation.]] | ||
== Conformational Changes == | == Conformational Changes == | ||
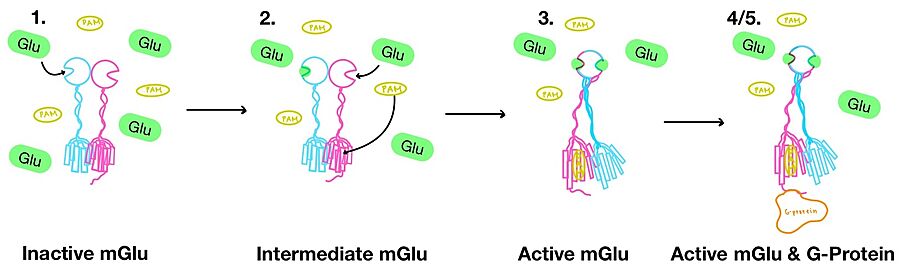
| - | '''1.''' | + | '''1.''' In its resting state, mGlu is in an <scene name='90/904320/Inactive_mglu2_first_picture/6'>inactive homodimeric form</scene> (Figure 3-1). In this conformation, the receptor is considered open with an inter-lobe angle of 44°<ref name="Seven">PMID:34194039</ref>. The structure has two free glutamate binding sites in the VFT, the CRDs are separated, and the TMD is not interacting with a G-protein<ref name="Seven">PMID:34194039</ref>. |
| - | '''2.''' | + | '''2.''' In the intermediate activation state, also known as the open-closed conformation, one glutamate is bound in one binding pocket of VFT (Figure 3-2). This single <scene name='90/904320/Mglu_binding/9'>glutamate bound state</scene> is still considered inactive as the receptor has not changed the conformations in the CRD and thus the TMD. With the same asymmetric transmembrane helices formation, a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> is still present and mGlu cannot interact with a G-protein<ref name="Seven">PMID:34194039</ref>. |
[[Image:Overview_mGlu_2.jpg|900 px|center|thumb|Figure 4. Illustration of mGlu's conformational change process.]] | [[Image:Overview_mGlu_2.jpg|900 px|center|thumb|Figure 4. Illustration of mGlu's conformational change process.]] | ||
| - | '''3.''' | + | '''3.''' A second glutamate then binds to the other <scene name='90/904320/Active_site_interactions/4'>binding pocket</scene> of the VFT (Figure 3-3). Mediated by L639, F643, N735, W773, and F776, a <scene name='90/904320/Pam/8'>positive allosteric modulator</scene> (PAM) also binds within the seven TMD helices of the alpha chain <ref name="Seven">PMID:34194039</ref>. This closed conformation of the VFT now has an inter-lobe angle of 25° is considered to be in the <scene name='90/904320/Active_mglu/10'>active conformation</scene><ref name="Seven">PMID:34194039</ref>. The binding of these [https://en.wikipedia.org/wiki/Ligand ligands] allows the CRDs to compact and come together. This conformational transformation causes the TMD to form a separate, active asymmetric conformation with a <scene name='90/904320/Active_helices/14'>TM6-TM6 interface</scene> between the chains (Figure 3)<ref name="Seven">PMID:34194039</ref>. |
[[Image:Screen Shot 2022-04-18 at 10.20.26 PM.png|300 px|right|thumb|Figure 5. The interaction between an active mGlu (magenta/lime/purple/crimson) and a G-protein (orange). Hydrogen bonds are shown through black dashes]] | [[Image:Screen Shot 2022-04-18 at 10.20.26 PM.png|300 px|right|thumb|Figure 5. The interaction between an active mGlu (magenta/lime/purple/crimson) and a G-protein (orange). Hydrogen bonds are shown through black dashes]] | ||
| - | '''4.''' | + | '''4.''' The crossover of the helices from the alpha and beta chains allows for intracellular loop 2 (ICL2) and the C-terminus to be properly ordered to interact with a single G-protein (Figure 3-4/5)<ref name="Seven">PMID:34194039</ref>. While hydrogen bonding is present between the C-terminus and alpha helix 5 of the G-protein, this <scene name='90/904320/Active_mglu/9'>mGlu/G-protein coupling</scene> is primarily driven by the hydrophobic interactions in the interface with the ɑ5 helix of the G-protein (Figure 5)<ref name="Seven">PMID:34194039</ref>. This coupling can only occur in the presence of a <scene name='90/904320/Pam/8'>PAM</scene> as the pocket in which the coupling occurs would be completely closed in its absence<ref name="Seven">PMID:34194039</ref>. |
| - | '''5.''' | + | '''5.''' Upon binding, the G-protein can become active through the receptor catalyzed reaction of GDP to GTP on the alpha subunit of the G-protein (Figure 3-4/5). Depending on the type of mGlu present, this activation causes different signaling cascades to occur within the cell <ref name="Lin">PMID:34135510</ref>. These cascades are necessary for cellular function as they can play primary roles in regulating metabolic molecules, [https://en.wikipedia.org/wiki/Ion_channel ion channels], transporter molecules, and several other parts of the cell; if these proteins are mutated, various diseases can occur<ref name="Crupi">PMID:30800054</ref>. |
== Clinical Relevance == | == Clinical Relevance == | ||
| - | Glutamate is a major neurotransmitter in the brain and is known for its role in memory, synaptic plasticity, and neuronal development<ref name="Crupi">PMID:30800054</ref>. Thus, mGlus are located throughout the CNS and play key roles in various neurodegenerative diseases such as [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's Disease] (AD), [https://en.wikipedia.org/wiki/Huntington%27s_disease Huntington's Disease] (HD), [https://en.wikipedia.org/wiki/Parkinson%27s_disease Parkinson’s Disease] (PD), and [https://en.wikipedia.org/wiki/Amyotrophic_lateral_sclerosis Amyotrophic Lateral Sclerosis] (ALS)<ref name="Crupi">PMID:30800054</ref>. Also, due to the key role of glutamate in the brain, mGlus are also being studied for their role in psychiatric disorders like [https://en.wikipedia.org/wiki/Anxiety anxiety] and [https://en.wikipedia.org/wiki/Depression_(mood) depression]<ref name="Crupi">PMID:30800054</ref>. In many of these diseases, the overexpression of glutamate can lead to the overstimulation of mGlus and excitotoxicity <ref name="Bordi">PMID:10416961</ref>. However, an exception to this is the under stimulation of mGlus which can result in schizophrenia<ref name="Conn">PMID:19058862</ref>. Research is ongoing, but studies have proven that agonists, antagonists, PAMs, and NAMs of mGlus are potential treatments for these diseases<ref name="Crupi">PMID:30800054</ref>. | + | Glutamate is a major neurotransmitter in the brain and is known for its role in memory, synaptic plasticity, and neuronal development<ref name="Crupi">PMID:30800054</ref>. Thus, mGlus are located throughout the CNS and play key roles in various neurodegenerative diseases such as [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's Disease] (AD), [https://en.wikipedia.org/wiki/Huntington%27s_disease Huntington's Disease] (HD), [https://en.wikipedia.org/wiki/Parkinson%27s_disease Parkinson’s Disease] (PD), and [https://en.wikipedia.org/wiki/Amyotrophic_lateral_sclerosis Amyotrophic Lateral Sclerosis] (ALS)<ref name="Crupi">PMID:30800054</ref>. Also, due to the key role of glutamate in the brain, mGlus are also being studied for their role in psychiatric disorders like [https://en.wikipedia.org/wiki/Anxiety anxiety] and [https://en.wikipedia.org/wiki/Depression_(mood) depression]<ref name="Crupi">PMID:30800054</ref>. In many of these diseases, the overexpression of glutamate can lead to the overstimulation of mGlus and [https://en.wikipedia.org/wiki/Excitotoxicity excitotoxicity] <ref name="Bordi">PMID:10416961</ref>. However, an exception to this is the under stimulation of mGlus which can result in [https://en.wikipedia.org/wiki/Schizophrenia schizophrenia]<ref name="Conn">PMID:19058862</ref>. Research is ongoing, but studies have proven that agonists, antagonists, PAMs, and NAMs of mGlus are potential treatments for these diseases<ref name="Crupi">PMID:30800054</ref>. |
== 3D Structures == | == 3D Structures == | ||
| Line 66: | Line 66: | ||
[[7mtr]], mGlu Active <br /> | [[7mtr]], mGlu Active <br /> | ||
[[7epb]], mGlu Active <br /> | [[7epb]], mGlu Active <br /> | ||
| - | [[7mts]], mGlu Active G Protein Bound <br /> | + | [[7mts]], mGlu Active G-Protein Bound <br /> |
== References == | == References == | ||
Current revision
Metabotropic Glutamate Receptor
| |||||||||||
Student Contributors
- Courtney Vennekotter
- Cade Chezem