Sandbox Reserved 1715
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 37: | Line 37: | ||
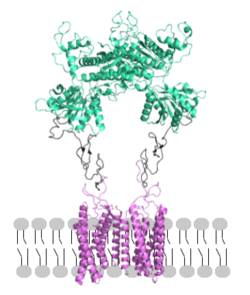
[[Image: Orientation_2.png|250px|left|thumb|Figure 2. Proper orientation of mGlu about the cell membrane.]] | [[Image: Orientation_2.png|250px|left|thumb|Figure 2. Proper orientation of mGlu about the cell membrane.]] | ||
==== Domains ==== | ==== Domains ==== | ||
| - | <scene name='90/904320/Mglu2_domains_vft/4'>VFT</scene>: The extracellular location in which the two glutamate agonists bind is known as the VFT. This domain includes a [https://en.wikipedia.org/wiki/Disulfide disulfide] bond between C121 of the alpha and beta chains <ref name="Du">PMID:34135509</ref>. This <scene name='90/904320/Inactive_mglu/ | + | <scene name='90/904320/Mglu2_domains_vft/4'>VFT</scene>: The extracellular location in which the two glutamate agonists bind is known as the VFT. This domain includes a [https://en.wikipedia.org/wiki/Disulfide disulfide] bond between C121 of the alpha and beta chains <ref name="Du">PMID:34135509</ref>. This <scene name='90/904320/Inactive_mglu/14'>disulfide bond</scene> is shifted down in the open conformation and undergoes a <scene name='90/904320/Cys_active/3'>disulfide bond upward movement</scene> upon glutamate binding which stabilizes the <scene name='90/904319/Vft/2'>active site</scene><ref name="Du">PMID:34135509</ref>. Glutamate binds within the <scene name='90/904320/Active_site_interactions/4'>binding pocket</scene> via [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding], specifically hydrogen bonding, with R57, S143, S145, T168, and K377 of the VFT<ref name="Seven">PMID:34194039</ref>. This binding initiates a closed VFT conformation. |
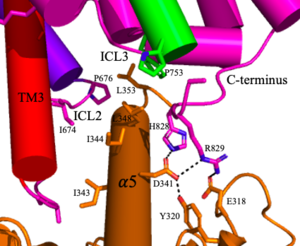
<scene name='90/904320/Mglu2_domains_crd/7'>CRD</scene>: The portion of the [https://en.wikipedia.org/wiki/Protomer protomer] that connects the VFT with the TMD is known as the CRD. Many <scene name='90/904320/Crd_cysteine/3'>disulfide bonds</scene> are located in this region between [https://en.wikipedia.org/wiki/Cysteine cysteines]. As the connecting segment of the protein, it is critical in transmitting the conformational change caused by the binding of glutamate in the VFT to the TMD<ref name="Seven">PMID:34194039</ref>. The change resulting from the binding of glutamate brings the cysteine-rich domains of the alpha and beta chain together to alter the configuration of the seven TMD helices through its interaction with the VFT extracellular loop 2 (ECL2) <ref name="Seven">PMID:34194039</ref>. This <scene name='90/904320/Active_helices/13'>ECL2 conformational change</scene> is mediated through interactions with amino acids at the apex of the CRD (e.g. I674, P676, and P753) <ref name="Seven">PMID:34194039</ref>. | <scene name='90/904320/Mglu2_domains_crd/7'>CRD</scene>: The portion of the [https://en.wikipedia.org/wiki/Protomer protomer] that connects the VFT with the TMD is known as the CRD. Many <scene name='90/904320/Crd_cysteine/3'>disulfide bonds</scene> are located in this region between [https://en.wikipedia.org/wiki/Cysteine cysteines]. As the connecting segment of the protein, it is critical in transmitting the conformational change caused by the binding of glutamate in the VFT to the TMD<ref name="Seven">PMID:34194039</ref>. The change resulting from the binding of glutamate brings the cysteine-rich domains of the alpha and beta chain together to alter the configuration of the seven TMD helices through its interaction with the VFT extracellular loop 2 (ECL2) <ref name="Seven">PMID:34194039</ref>. This <scene name='90/904320/Active_helices/13'>ECL2 conformational change</scene> is mediated through interactions with amino acids at the apex of the CRD (e.g. I674, P676, and P753) <ref name="Seven">PMID:34194039</ref>. | ||
| Line 45: | Line 45: | ||
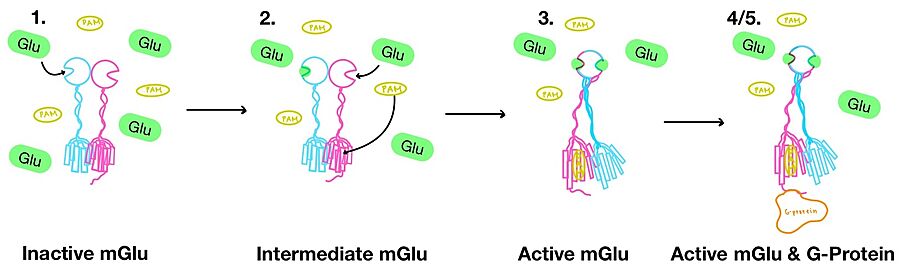
== Conformational Changes == | == Conformational Changes == | ||
| - | '''1.''' In its resting state, mGlu is in an <scene name='90/904320/Inactive_mglu2_first_picture/6'>inactive homodimeric form</scene>(Figure 3-1). In this conformation, the receptor is considered open with an inter-lobe angle of 44°<ref name="Seven">PMID:34194039</ref>. The structure has two free glutamate binding sites in the VFT, the CRDs are separated, and the TMD is not interacting with a G-protein<ref name="Seven">PMID:34194039</ref>. | + | '''1.''' In its resting state, mGlu is in an <scene name='90/904320/Inactive_mglu2_first_picture/6'>inactive homodimeric form</scene> (Figure 3-1). In this conformation, the receptor is considered open with an inter-lobe angle of 44°<ref name="Seven">PMID:34194039</ref>. The structure has two free glutamate binding sites in the VFT, the CRDs are separated, and the TMD is not interacting with a G-protein<ref name="Seven">PMID:34194039</ref>. |
'''2.''' In the intermediate activation state, also known as the open-closed conformation, one glutamate is bound in one binding pocket of VFT (Figure 3-2). This single <scene name='90/904320/Mglu_binding/9'>glutamate bound state</scene> is still considered inactive as the receptor has not changed the conformations in the CRD and thus the TMD. With the same asymmetric transmembrane helices formation, a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> is still present and mGlu cannot interact with a G-protein<ref name="Seven">PMID:34194039</ref>. | '''2.''' In the intermediate activation state, also known as the open-closed conformation, one glutamate is bound in one binding pocket of VFT (Figure 3-2). This single <scene name='90/904320/Mglu_binding/9'>glutamate bound state</scene> is still considered inactive as the receptor has not changed the conformations in the CRD and thus the TMD. With the same asymmetric transmembrane helices formation, a <scene name='90/904320/Inactive_tmd_interface/1'>TM3-TM4 interface</scene> is still present and mGlu cannot interact with a G-protein<ref name="Seven">PMID:34194039</ref>. | ||
Current revision
Metabotropic Glutamate Receptor
| |||||||||||
Student Contributors
- Courtney Vennekotter
- Cade Chezem