Sandbox Reserved 1716
From Proteopedia
(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 9: | Line 9: | ||
Vitamin K Epoxide Reductase is found and primarily synthesized in the [https://en.wikipedia.org/wiki/Liver liver] . | Vitamin K Epoxide Reductase is found and primarily synthesized in the [https://en.wikipedia.org/wiki/Liver liver] . | ||
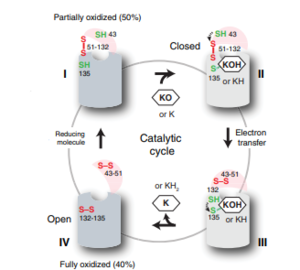
| - | Vitamin K Epoxide Reductase is unstable in-vitro. To determine its structure an extra protein superfolder green flourescent protein (sGFP) was appended to the N and C termini of Vitamin K Epoxide<ref name= | + | Vitamin K Epoxide Reductase is unstable in-vitro. To determine its structure an extra protein superfolder green flourescent protein (sGFP) was appended to the N and C termini of Vitamin K Epoxide<ref name="Liu">PMID:33154105</ref> . For the visualizing VKOR, this protein has been removed from the structural scenes. After sGFP was removed from the structural scenes, we further took the structure files and resequenced them to better align with the numbering of the protein. In these files the sequence slightly differs between the organisms used to view Vitamin K Epoxide Reductase. In the human version, HsVKOR, the catalytic cysteines that play an intricate role in the reduction of Vitamin K Epoxide are <scene name='90/904321/Cysteines/7'>cysteines</scene> 43, 51, 132, and 135. In the pufferfish version of the file, TrVKORL, the cysteines are 52, 55, 141, and 144. |
| Line 15: | Line 15: | ||
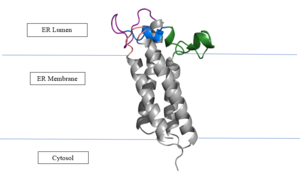
[[Image:VKORmembrane.PNG|300px|right|thumb|'''Figure 2: Vitamin K Epoxide Reductase in the Endoplasmic Membrane''']] | [[Image:VKORmembrane.PNG|300px|right|thumb|'''Figure 2: Vitamin K Epoxide Reductase in the Endoplasmic Membrane''']] | ||
| - | In the liver, the VKOR enzyme is set in the endoplasmic reticulum membrane (Fig.2). The transmembrane helices are located in the Endoplasmic Reticulum Luminal Region, which is the region between the ER Lumen and the [https://en.wikipedia.org/wiki/Cytosol Cytosol]. The cap region is partially oriented in the ER Lumen. The active site remains within the [https://en.wikipedia.org/wiki/Endoplasmic_reticulum Endoplasmic Reticulum Membrane].The Anchor is partially within the ER lumen, and partially embedded in the ER membrane. The anchor is what attaches the cap domain and stabilizes it, which allows the cap domain to cover the active site.<ref name="Liu" | + | In the liver, the VKOR enzyme is set in the endoplasmic reticulum membrane (Fig.2). The transmembrane helices are located in the Endoplasmic Reticulum Luminal Region, which is the region between the ER Lumen and the [https://en.wikipedia.org/wiki/Cytosol Cytosol]. The cap region is partially oriented in the ER Lumen. The active site remains within the [https://en.wikipedia.org/wiki/Endoplasmic_reticulum Endoplasmic Reticulum Membrane].The Anchor is partially within the ER lumen, and partially embedded in the ER membrane. The anchor is what attaches the cap domain and stabilizes it, which allows the cap domain to cover the active site.<ref name="Liu"/> |
The VKOR enzyme is made up of four transmembrane helices (Fig.2): <scene name='90/904321/Tm1/2'>TM1</scene>, <scene name='90/904321/Tm2/3'>TM2</scene>, <scene name='90/904321/Tm3/3'>TM3</scene>, and <scene name='90/904321/Tm4/3'>TM4 </scene>(Grey/Orange). Each of these helices come together to form a central ligand binding pocket. This central pocket is the active site where conserved Cysteines: C132 and C135 are located. In the cap domain are important regions that are significant for Vitamin K binding, and the overall function of Vitamin K Epoxide Reductase, including the Anchor(Green), Cap Sequence (Blue), Beta Hairpin (Purple), and 3-4 Loop (Pink). <ref name="Liu"/> | The VKOR enzyme is made up of four transmembrane helices (Fig.2): <scene name='90/904321/Tm1/2'>TM1</scene>, <scene name='90/904321/Tm2/3'>TM2</scene>, <scene name='90/904321/Tm3/3'>TM3</scene>, and <scene name='90/904321/Tm4/3'>TM4 </scene>(Grey/Orange). Each of these helices come together to form a central ligand binding pocket. This central pocket is the active site where conserved Cysteines: C132 and C135 are located. In the cap domain are important regions that are significant for Vitamin K binding, and the overall function of Vitamin K Epoxide Reductase, including the Anchor(Green), Cap Sequence (Blue), Beta Hairpin (Purple), and 3-4 Loop (Pink). <ref name="Liu"/> | ||
| Line 73: | Line 73: | ||
Warfarin still forms Hydrogen bonds with <scene name='90/904322/Asn80_tyr139_warfarin/1'>Asn80 and Tyr139</scene>. The specific bonds are between Asn80 and the 2-ketone group of warfarin and Tyr139 with the 4-hydroxyl group of warfarin. The rest of the pocket is hydrophobic interactions. The H bonds are necessary for the recognition of the ligand in the binding site of VKOR. | Warfarin still forms Hydrogen bonds with <scene name='90/904322/Asn80_tyr139_warfarin/1'>Asn80 and Tyr139</scene>. The specific bonds are between Asn80 and the 2-ketone group of warfarin and Tyr139 with the 4-hydroxyl group of warfarin. The rest of the pocket is hydrophobic interactions. The H bonds are necessary for the recognition of the ligand in the binding site of VKOR. | ||
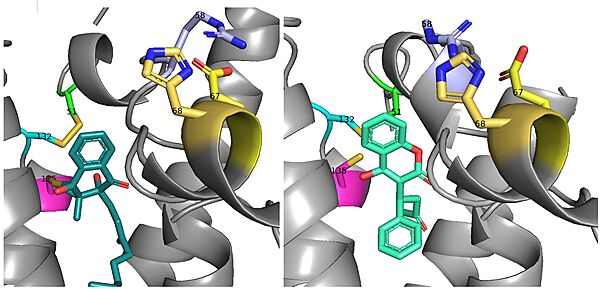
| - | There is a slight difference in the way in which warfarin binds compared to VKO. Warfarin binds | + | There is a slight difference in the way in which warfarin binds compared to VKO. Warfarin binds at a 30 degree angle from where VKO would bind (Fig.8). This creates a difference in how the cap loop and anchor domain interact, and that noticeable difference is with <scene name='90/904322/Arg58/4'>Arg58</scene>. With VKO, Arg58, located in the cap loop, directly interacts with <scene name='90/904322/Arg58_vko/6'>Glu67</scene> when VKO is bound. When warfarin binds, Arg58 is found inserted between <scene name='90/904322/Arg58_warfarin/3'>Glu67 and His68</scene> of the anchor domain (Fig. 8).<ref name="Liu"/> |
[[Image:VKO and Warfarin binding.jpg|600 px|right|thumb|'''Figure 8. Vitamin K Epoxide and Warfarin Binding Angles:''' There is a slight angle of around 30 degrees in which VKO(left) and warfarin(right) bind. The location of the cap domain and the interactions between amino acid residues changes based on this slight difference in binding.]] | [[Image:VKO and Warfarin binding.jpg|600 px|right|thumb|'''Figure 8. Vitamin K Epoxide and Warfarin Binding Angles:''' There is a slight angle of around 30 degrees in which VKO(left) and warfarin(right) bind. The location of the cap domain and the interactions between amino acid residues changes based on this slight difference in binding.]] | ||
Current revision
Vitamin K Epoxide Reductase
| |||||||||||