Sphingosine kinase
From Proteopedia
(Difference between revisions)
| (28 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
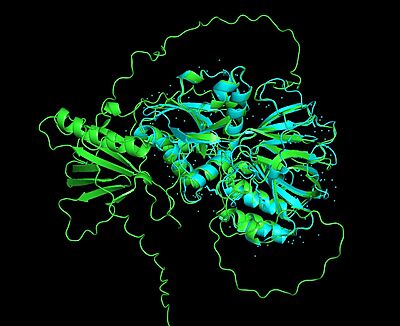
| - | <StructureSection load='3vzb' size='350' side='right' caption=' | + | <StructureSection load='3vzb' size='350' side='right' caption='Human sphingosine kinase, Type 1 complex with sphingosine, ethylene glycol and sulphate (PDB entry [[3vzb]])' scene=''> |
| + | __TOC__ | ||
==Structure Highlights== | ==Structure Highlights== | ||
<table><tr><td colspan='2'> For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3VZB FirstGlance]. <br> | <table><tr><td colspan='2'> For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3VZB FirstGlance]. <br> | ||
| Line 6: | Line 7: | ||
<tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">SPHK1, SPHK, SPK ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 HUMAN])</td></tr> | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">SPHK1, SPHK, SPK ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=9606 HUMAN])</td></tr> | ||
<tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Sphinganine_kinase Sphinganine kinase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.7.1.91 2.7.1.91] </span></td></tr> | <tr id='activity'><td class="sblockLbl"><b>Activity:</b></td><td class="sblockDat"><span class='plainlinks'>[http://en.wikipedia.org/wiki/Sphinganine_kinase Sphinganine kinase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.7.1.91 2.7.1.91] </span></td></tr> | ||
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3vzb FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3vzb OCA | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3vzb FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3vzb OCA], [http://www.rcsb.org/pdb/explore.do?structureId=3vzb RCSB]</span></td></tr> |
| + | |||
</table> | </table> | ||
| Line 13: | Line 15: | ||
==Structure Overview== | ==Structure Overview== | ||
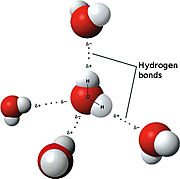
| - | [[Image:3D hydrogen Bonding.jpg |thumb|right| Figure 1. Hydrogen Bonding Schematic]] | + | [[Image:3D hydrogen Bonding.jpg |thumb|right| '''Figure 1'''. Hydrogen Bonding Schematic]] |
The '''Sphingosine Kinase Type 1 (SphK1)''' structure reveals a two domain '''alpha-beta''' architecture that comprises <scene name='91/910628/Alpha_hel_and_b_sheets_of_sphk/1'>9 α helices and 17 β strands</scene>.<ref name="wang">PMID:23602659</ref> Particularly, the N-terminal domain (NTD) adopts an α/β fold and comprises 6 α helices and 6 β strands, resembling the dinucleotide binding motif of a [[Rossmann fold]] of a three-layer α/β/α sandwich (discussed further in the "Sphingosine Kinases are Evolutionarily Conserved" section below). <ref name="wang">PMID:23602659</ref> Alternatively, the C-terminal domain (CTD) comprises 11 β strands and 4 α helices. Particularly, the 11 β strands form an antiparallel β sandwich. <ref name="wang">PMID:23602659</ref> | The '''Sphingosine Kinase Type 1 (SphK1)''' structure reveals a two domain '''alpha-beta''' architecture that comprises <scene name='91/910628/Alpha_hel_and_b_sheets_of_sphk/1'>9 α helices and 17 β strands</scene>.<ref name="wang">PMID:23602659</ref> Particularly, the N-terminal domain (NTD) adopts an α/β fold and comprises 6 α helices and 6 β strands, resembling the dinucleotide binding motif of a [[Rossmann fold]] of a three-layer α/β/α sandwich (discussed further in the "Sphingosine Kinases are Evolutionarily Conserved" section below). <ref name="wang">PMID:23602659</ref> Alternatively, the C-terminal domain (CTD) comprises 11 β strands and 4 α helices. Particularly, the 11 β strands form an antiparallel β sandwich. <ref name="wang">PMID:23602659</ref> | ||
| - | The <scene name='91/910628/Sphk1_surface/4'>catalytic site</scene> (used in converting sphingosine to S1P-see below in "Sphingolipids" section for more details) is in a <scene name='91/910628/Sphk1_hydrophobic_pocket/1'>'''hydrophobic''' cleft</scene>, located between the two domains. <ref name="wang">PMID:23602659</ref> This hydrophobic cleft is an '''ideal binding pocket''' for a '''hydrophobic lipid''', like sphingosine (an '''18-carbon''' amino alcohol with an '''unsaturated''' '''hydrocarbon''' chain). <ref name="wang">PMID:23602659</ref> The binding of sphingosine to the SphK1 hydrophobic "pocket" is mediated by both the anchoring of the <scene name='91/910628/Sphingosine_element_label/2'>hydrophilic (polar) headgroup</scene> of sphingosine to the protein surface and the presence of the hydrophobic alkyl chain in the interior of the protein. <ref name="wang">PMID:23602659</ref> The 2-amino-1,3-diol moiety of the sphingosine head group is situated at the cleft between the two domains, allowing for '''hydrogen-bond interactions''' | + | The <scene name='91/910628/Sphk1_surface/4'>catalytic site</scene> (used in converting sphingosine to S1P-see below in "Sphingolipids" section for more details) is in a <scene name='91/910628/Sphk1_hydrophobic_pocket/1'>'''hydrophobic''' cleft</scene>, located between the two domains. <ref name="wang">PMID:23602659</ref> This hydrophobic cleft is an '''ideal binding pocket''' for a '''hydrophobic lipid''', like sphingosine (an '''18-carbon''' amino alcohol with an '''unsaturated''' '''hydrocarbon''' chain). <ref name="wang">PMID:23602659</ref> The binding of sphingosine to the SphK1 hydrophobic "pocket" is mediated by both the anchoring of the <scene name='91/910628/Sphingosine_element_label/2'>hydrophilic (polar) headgroup</scene> of sphingosine to the protein surface and the presence of the hydrophobic alkyl chain in the interior of the protein. <ref name="wang">PMID:23602659</ref> The 2-amino-1,3-diol moiety of the sphingosine head group is situated at the cleft between the two domains, allowing for '''hydrogen-bond interactions''' ('''Figure 1''') with <scene name='91/910628/Amino_acid_interactions/4'>Asp81 through the 1-hydroxyl and with Asp178 through the 3-hydroxyl</scene>, which also forms a water-mediated hydrogen bond with Ser168. Sphingosine's long acyl chain is buried in the hydrophobic pocket ("tunnel shaped"), lined by side chains of mostly <scene name='91/910628/Amino_acid_interactions/6'>non-polar amino acid residues</scene> (i.e., Phe173, Ile174, Val177, Phe192, Thr196, Leu299, Leu302, Phe303, Met306, His311, Leu319, Phe288, Val290, Leu259, Leu261, Ala170, Met272, and Ala274). <ref name="wang">PMID:23602659</ref> |
| - | + | ||
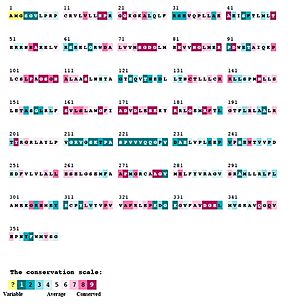
| - | [[Image:SphK1 sequence conservation Large.jpeg| | + | [[Image:SphK1 Sequence and Structure Conservation Small.jpeg|300px|right|thumb|'''Figure 2'''. SphK1 Sequence Conservation Structure with Conservation Table in Figure 4]] |
| + | [[Image:SphK1 sequence conservation Large.jpeg|300px|right|thumb|'''Figure 3'''. SphK1 Sequence Conservation with Conservation Scale]] | ||
==Sphingosine Kinases are Evolutionarily Conserved== | ==Sphingosine Kinases are Evolutionarily Conserved== | ||
| - | [https://en.wikipedia.org/wiki/Kinase '''Sphingosine Kinases'''] are part of distinct class of lipid kinases that is evolutionarily highly conserved.<ref name="Kohama">PMID:9726979</ref> They do not share sequence homology with other lipid kinases, such as phosphotidylinositol-3 kinase [https://en.wikipedia.org/wiki/Phosphoinositide_3-kinase (PI3K)]. <ref name="wang">PMID:23602659</ref> However, protein sequence motif analysis and structure classification suggested that SphKs belong to the phosphofructokinase [https://en.wikipedia.org/wiki/Phosphofructokinase (PFK)]-like superfamily <ref name="cheek">PMID:15771780</ref> <ref name="labesse">PMID:12069781</ref> sharing the same protein fold (i.e. the [[Rossmann fold]]) with NAD kinases, ceramide kinases, and phosphofructokinases. <ref name="wang">PMID:23602659</ref> | + | [https://en.wikipedia.org/wiki/Kinase '''Sphingosine Kinases'''] are part of distinct class of lipid kinases that is evolutionarily highly conserved ('''Figure 2 and 3''').<ref name="Kohama">PMID:9726979</ref> They do not share sequence homology with other lipid kinases, such as phosphotidylinositol-3 kinase [https://en.wikipedia.org/wiki/Phosphoinositide_3-kinase (PI3K)]('''Figure 3'''). <ref name="wang">PMID:23602659</ref> However, protein sequence motif analysis and structure classification suggested that SphKs belong to the phosphofructokinase [https://en.wikipedia.org/wiki/Phosphofructokinase (PFK)]-like superfamily <ref name="cheek">PMID:15771780</ref> <ref name="labesse">PMID:12069781</ref> sharing the same protein fold (i.e. the [[Rossmann fold]]) with NAD kinases, ceramide kinases, and phosphofructokinases. <ref name="wang">PMID:23602659</ref> |
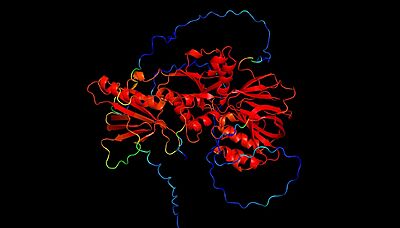
| - | The mammalian isoforms of sphingosine kinase, SphK1 and SphK2, are the only isoforms that have been characterized. Additionally, the crystal structure has been solved for SphK1 (via [[X-ray diffraction]] resulting in a resolution of 2.00Å) with SphK2 having a highly predictive [https://en.wikipedia.org/wiki/AlphaFold Alpha Fold] structure. <ref name="wang">PMID:23602659</ref> | + | The mammalian isoforms of sphingosine kinase, SphK1 and SphK2, are the only isoforms that have been characterized ('''Figure 4 and 5'''). Additionally, the crystal structure has been solved for SphK1 (via [[X-ray diffraction]] resulting in a resolution of 2.00Å) with SphK2 having a highly predictive [https://en.wikipedia.org/wiki/AlphaFold Alpha Fold] structure ('''Figure 4 and 5'''). <ref name="wang">PMID:23602659</ref> '''Figure 5''' shows the alignment of the crystal structure of SphK1 and the Alpha Fold prediction model of SphK2. Here, the catalytic domain aligns with high confidence (showing an RMSD value of 0.0846Å. However, the Alpha Fold prediction model of Sphk2 shows an entire domain (of α/β structure architecture) that does not align with the SphK1 crystal structure. As you will read further in the "Sphingosine Kinase (SphK1 and SphK2)" section, although SphK1 and SphK2 share similar structural properties and mechanistic properties, their kinetic regulation is different (a reason as to why there may be an additional domain predicted in the SphK2 structure). |
| - | [[Image:SphK2 AlphaFold B-factor.jpeg| | + | [[Image:SphK2 AlphaFold B-factor.jpeg|400px|left|thumb|'''Figure 4'''. SphK2 Alpha Fold Prediction Model, Labeled by B-factor (red=high confidence prediction, blue=low confidence prediction]] |
| - | [[Image:SphK1 and SphK2 alignment Large.jpeg|400px|center|thumb|Figure | + | [[Image:SphK1 and SphK2 alignment Large.jpeg|400px|center|thumb|'''Figure 5'''. SphK1 and SphK2 Alignment, RMSD= 0.846Å, SphK1=cyan, SphK2=lime green]] |
== Sphingolipids == | == Sphingolipids == | ||
[https://en.wikipedia.org/wiki/Sphingolipid '''Sphingolipids'''] are a class of lipids, containing a [https://en.wikipedia.org/wiki/Sphingosine#:~:text=Sphingosine%20(2%2Damino%2D4,include%20sphingomyelin%2C%20an%20important%20phospholipid '''sphingosine'''] base moiety, essential for eukaryotic cell membrane structure and function. <ref name="futerman">PMID:15289826</ref> Importantly, sphingolipids can additionally act as critical signaling molecules used in many eukaryotic homeostatic cellular processes, such as inflammation, proliferation, apoptosis, cell migration, and '''pathogen defense'''. <ref name="futerman">PMID:15289826</ref> Because of this, it is critical for sphingolipids and their metabolizing enzymes to exist in a delicate homeostatic balance within the eukaryotic cell system. | [https://en.wikipedia.org/wiki/Sphingolipid '''Sphingolipids'''] are a class of lipids, containing a [https://en.wikipedia.org/wiki/Sphingosine#:~:text=Sphingosine%20(2%2Damino%2D4,include%20sphingomyelin%2C%20an%20important%20phospholipid '''sphingosine'''] base moiety, essential for eukaryotic cell membrane structure and function. <ref name="futerman">PMID:15289826</ref> Importantly, sphingolipids can additionally act as critical signaling molecules used in many eukaryotic homeostatic cellular processes, such as inflammation, proliferation, apoptosis, cell migration, and '''pathogen defense'''. <ref name="futerman">PMID:15289826</ref> Because of this, it is critical for sphingolipids and their metabolizing enzymes to exist in a delicate homeostatic balance within the eukaryotic cell system. | ||
| - | [https://en.wikipedia.org/wiki/Sphingosine-1-phosphate '''Sphingosine-1-Phosphate (S1P)'''], a lipid within the sphingolipid class, is an important signaling molecule that can participate in intracellular and extracellular signaling. <ref name="futerman">PMID:15289826</ref> <ref name="Spiegel">PMID:12728273</ref> If meant to be an intracellular signaling molecule, S1P is important for cell survival, growth, and overall cell function.<ref name="Spiegel">PMID:12728273</ref> Alternatively, if S1P is meant to be an extracellular signaling molecule, later binding to one of five different [https://en.wikipedia.org/wiki/Sphingosine-1-phosphate_receptor S1P G-Protein Coupled Receptors] on the same or different cell type, it is used to activate many signaling cascades important for | + | [https://en.wikipedia.org/wiki/Sphingosine-1-phosphate '''Sphingosine-1-Phosphate (S1P)'''], a lipid within the sphingolipid class, is an important signaling molecule that can participate in intracellular and extracellular signaling. <ref name="futerman">PMID:15289826</ref> <ref name="Spiegel">PMID:12728273</ref> If meant to be an intracellular signaling molecule, S1P is important for cell survival, growth, and overall cell function.<ref name="Spiegel">PMID:12728273</ref> Alternatively, if S1P is meant to be an extracellular signaling molecule, later binding to one of five different [https://en.wikipedia.org/wiki/Sphingosine-1-phosphate_receptor S1P G-Protein Coupled Receptors] on the same or different cell type, it is used to activate many signaling cascades important for cellular response. <ref name="Spiegel">PMID:12728273</ref> One '''important function of S1P''', whether its fate serves intracellularly or extracellularly, is '''eliciting the [https://en.wikipedia.org/wiki/Immune_response immune response] (i.e. [https://en.wikipedia.org/wiki/Lymphocyte lymphocyte] trafficking)''', especially as a mechanism in [https://en.wikipedia.org/wiki/Pathogen '''pathogen] defense''' (please see section below labeled "Importance of SphKs in Pathogen Defense and Immune Response" for more information about SphKs involvement within the immune response). <ref name="Spiegel">PMID:12728273</ref> |
== Sphingosine Kinase (SphK1 and SphK2)== | == Sphingosine Kinase (SphK1 and SphK2)== | ||
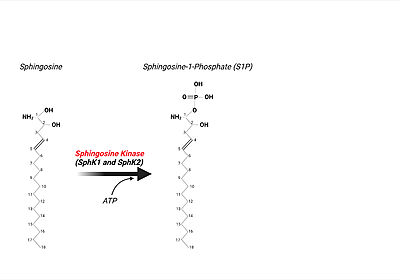
| - | [[Image:Sphingosine to S1P.jpeg|400px|right|thumb|The Phosphorylation of Sphingosine by SphK]] | + | [[Image:Sphingosine to S1P.jpeg|400px|right|thumb|'''Figure 6'''. The Phosphorylation of Sphingosine by SphK]] |
| - | '''Sphingosine Kinase''', existing in two isoenzyme forms, SphK1 and SphK2, '''creates S1P via the phosphorylation of sphingosine''', the base moiety of all sphingolipids. <ref name="Spiegel">PMID:12728273</ref> Particularly, this catalytic reaction is [[ATP]] ('''a'''denine''' t'''ri'''p'''hosphate)-dependent | + | '''Sphingosine Kinase''', existing in two isoenzyme forms, SphK1 and SphK2, '''creates S1P via the phosphorylation of sphingosine''', the base moiety of all sphingolipids ('''Figure 6'''). <ref name="Spiegel">PMID:12728273</ref> Particularly, this catalytic reaction is [[ATP]] ('''a'''denine''' t'''ri'''p'''hosphate)-dependent aiding the phosphorylation of sphingosine on the primary hydroxyl group. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | Sphingosine Kinase(s) are a key enzyme controlling the levels of S1P within the eukaryotic cell, and thus, is an important regulator of diverse cellular functions (as mentioned previously). <ref name="wang">PMID:23602659</ref> As there are'''two isoforms''' of sphingosine kinase, '''SphK1''' and '''SphK2''', each isoform has it own specificities. <ref name="Drexler">PMID:15289826</ref> For instance, SphK1 and SphK2 are '''tissue specific'''. SphK1 is prevalent in the lung and spleen, whereas SphK2 is prevalent in the liver and heart. <ref name="Spiegel">PMID:12728273</ref> Importantly, although SphK1 and SphK2 catalyze the same biochemical reactions, they originate from different genes and have different substrate specificities, tissue distributions (like mentioned above), and subcellular localization patterns.<ref name="wang">PMID:23602659</ref> Typically sphingolipid metabolizing enzymes are located along the [https://en.wikipedia.org/wiki/Endoplasmic_reticulum Endoplasmic Reticulum (ER)]. <ref name="Spiegel">PMID:12728273</ref> Particularly, SphK2 has 4 transmembrane domains and is thus localized to the membrane of the ER where it serves to phosphorylate circulating sphingosine when signaled to do so. <ref name="Spiegel">PMID:12728273</ref> Alternatively, SphK1 has '''no''' transmembrane domains (being, instead, located within the [https://en.wikipedia.org/wiki/Cytoplasm cytoplasm] close to the ER organelle). <ref name="Spiegel">PMID:12728273</ref> However, it is unknown whether SphK1 is tethered by some other molecule to the ER or remains untethered within the cytoplasmic compartment. Although SphK1 and SphK2 are similar in function, they are additionally diverse in the localization within the eukaryotic cell environment with differing kinetic properties (i.e., prevalent kinetic regulation differences). <ref name="Spiegel">PMID:12728273</ref> | ||
== Importance of SphKs in Pathogen Defense and Immune Response == | == Importance of SphKs in Pathogen Defense and Immune Response == | ||
| - | Upon the '''SphK ATP-dependent phosphorylation event of sphingosine to S1P''', the phosphorylated lipid product can be utilized in many intracellular and extracellular signaling mechanisms. <ref name="Drexler">PMID:15289826</ref> One '''critical''' function of S1P (through [https://en.wikipedia.org/wiki/Autocrine_signaling autocrine] or [https://en.wikipedia.org/wiki/Paracrine_signaling paracrine] extracellular signaling), is eliciting the immune response when pathogens are present. <ref name="Bryan">PMID:15289826</ref> | + | Upon the '''SphK ATP-dependent phosphorylation event of sphingosine to S1P''', the phosphorylated lipid product can be utilized in many intracellular and extracellular signaling mechanisms. <ref name="Drexler">PMID:15289826</ref> One '''critical''' function of S1P (through [https://en.wikipedia.org/wiki/Autocrine_signaling autocrine] or [https://en.wikipedia.org/wiki/Paracrine_signaling paracrine] extracellular signaling), is eliciting the immune response when pathogens are present or causing infection. <ref name="Bryan">PMID:15289826</ref> In order to "kick-start" the immune response, first, S1P is transported out of the cell by Spinster Homologue 2, SPNS2, a member of a large family of non ATP-dependent [https://en.wikipedia.org/wiki/Ion_transporter ion transporters]. <ref name="Spiegal2">PMID:30655317</ref> After the lipid's transport out of eukaryotic cell, S1P can bind to one of '''5 S1P G-protein coupled receptors (GPCRs)''' present on the surface of both innate and adaptive immune cell types resulting in immune cell trafficking to the site of infection. <ref name="Spiegal2">PMID:30655317</ref> <ref name="Bryan">PMID:15289826</ref> For example, [https://en.wikipedia.org/wiki/Macrophage macrophages] express S1P receptors I, II, III, and IV. Upon binding of S1P to S1P receptor type I, macrophages are signaled to be trafficked and recruited to the site of pathogen infection. <ref name="Bryan">PMID:15289826</ref> |
| + | ==3D structures of sphingosine kinase== | ||
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | [[4v24]] – hSphK1 kinase domain 1-363 + pyrrolidine derivative – human<br /> | ||
| + | [[3vzb]] – hSphK1 kinase domain + sphingosine <br /> | ||
| + | [[3vzc]], [[4l02]] – hSphK1 kinase domain + inhibitor <br /> | ||
| + | [[3vzd]] – hSphK1 kinase domain + inhibitor + ADP<br /> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | </StructureSection> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||