User:Arthur Migliatti/Sandbox1
From Proteopedia
< User:Arthur Migliatti(Difference between revisions)
| (68 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
== Introduction == | == Introduction == | ||
| - | This is a default text for your page '''Arthur Migliatti/Sandbox1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
<StructureSection load='1trs' size='340' side='right' caption='Human Thioredoxin 1'> | <StructureSection load='1trs' size='340' side='right' caption='Human Thioredoxin 1'> | ||
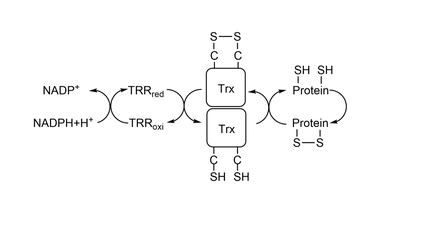
| - | '''Thioredoxin'''(Trx) is a protein present in all organisms, from bacterias to complex beings as humans. This page will be focused on exploring the characteristics of '''Trx1''', a cytosolic form of Trx present in eukaryotes. Trx1 has an active site composed of 2 cysteines separated by 2 aminoacids (<scene name='91/911850/Trx_cys-active_site/ | + | '''Thioredoxin'''(Trx) is a protein present in all organisms, from bacterias to complex beings as humans. This page will be focused on exploring the characteristics of '''Trx1''', a cytosolic form of Trx present in eukaryotes. Trx1 has an active site composed of 2 cysteines separated by 2 aminoacids (<scene name='91/911850/Trx_cys-active_site/2'>Cys32 - X - X - Cys35</scene>) which catalyses the reduction of other thiol-proteins and becomes oxidized. It is reduced back by '''[[Thioredoxin Reductase]]'''(TrxR), which, in the end, is reduced by '''NADPH'''. Together, the two proteins and NADPH form the system Trx<ref>Lu, J.; Holmgren, A. The Thioredoxin Antioxidant System. Free Radical Biology and Medicine 2014, 66, 75–87. https://doi.org/10.1016/j.freeradbiomed.2013.07.036.</ref><ref>Netto, L. E. S.; de Oliveira, M. A.; Tairum, C. A.; da Silva Neto, J. F. Conferring Specificity in Redox Pathways by Enzymatic Thiol/Disulfide Exchange Reactions. Free Radical Research 2016, 50 (2), 206–245. https://doi.org/10.3109/10715762.2015.1120864. |
| + | </ref>(Figure 1). As it is shown in this <scene name='91/911850/Conservation/1'>image</scene>, both Cys32 and Cys 35 were highly conserved during evolution(<font color='mediumvioletred'><b>dark pink</b></font> = highly conserved, <span style="color:white;background-color:black;font-weight:bold;">white</span> = average, <font color='deepskyblue'><b>blue</b></font> = variable). One of the most important proteins that Trx reduces is '''[[Peroxiredoxin]]'''(Prx), which catalyses the reduction of Hidrogen Peroxide(H2O2) to water. Since high concentrations of H2O2 produces other potent oxidizing molecules, such as hydroxyl radical, Prx's action, and so Trx's also, are fundamental for the cell to have a redox homeostasis and to have low amount of damage. | ||
| - | + | [[Image:ThioredoxinSystem.png|thumb|center|540x240px|Figure 1: Thioredoxin System.]] | |
| - | <scene name='91/911850/ | + | To reduce other proteins, first happens an attack from Cys32, creating an intermolecular disulfide bond, represented <scene name='91/911850/C32_s-s_c206/1'>here</scene> between residue Cys32 from Trx1 and residue Cys206 from '''[[MsrA]]'''.After it, residue Cys35 attacks Cys32, creating a <scene name='91/911850/Trx_cys_-_oxidized_-_diss_bond/4'>disulfide bond</scene> between the two cysteines in Trx1's catalytic site. To study the mechanism of reduction by Trx1 it is commom to use a wild-type Trx1 Cys35Ser. Since there is no other Cys to make a disulfide bond with Cys32, Trx1 is mainteined bonded to the other protein being studied by a intermolecular disulfide bond. |
| - | + | == Structure == | |
| - | <scene name='91/911850/ | + | Trx1 is a monomeric protein and weights around 12kDa. It is formed by one five-stranded beta sheet involved by 4 alpha helix, shown <scene name='91/911850/Secondary_structure/1'>here</scene>. The active site is located on a lump between betra strand 2, where Cys 35 is located, and alpha helix 2, where Cys32 is located. By being in the end of an alpha helix, <scene name='91/911850/Secondary_structure_cys32/1'>Cys32</scene> has a lower pKa, making it possible to reduce disulfide bonds<ref>Holmgren, A. Thioredoxin Structure and Mechanism: Conformational Changes on Oxidation of the Active-Site Sulfhydryls to a Disulfide. Structure 1995, 3 (3), 239–243. https://doi.org/10.1016/S0969-2126(01)00153-8.</ref>. As it is shown in this <scene name='91/911850/Dipolo_positivo_no_sitio_ativo/1'>image</scene>, the nitrogens point to the active site, while the oxigens point to the other direction. This creates a positive dipole, which reduces the pKa of the active site. |
| - | <scene name='91/911850/Trx-cys-red-dislig-s-ca/ | + | The structure of the protein doesn't change when it goes from reduced to oxidized, but the sulfur of the cysteines in the active site come closer, although both cysteines stay in the same distance(<scene name='91/911850/Trx-oxi-dislig-s-s_-_ca-ca/2'>oxidized</scene> x <scene name='91/911850/Trx-cys-red-dislig-s-ca/3'>reduced</scene>). As it's possible to see in the next images, the dihedral angle made by N - Calpha - C - S changes in both cysteines. In the <scene name='91/911850/Trx-cys-red-ang-s-ca/1'>reduced state</scene>, the angles are in such way that the sulfur atoms are far apart. When it is in the <scene name='91/911850/Trx-oxi-dislig-s-s_-_ca-ca/3'>oxidized state</scene>, the sulfur atoms come closer together and bond. |
| - | + | == Function == | |
| - | + | Trx and TrxR were first discovered in 1964 in a study realized in bacteria, and were described as necessary proteins to reduce '''[[Ribonucleotide Reductase]]'''(RNR), a protein that produces deoxyribonucleotides from ribonucleotides<ref>Laurent, T. C.; Moore, E. C.; Reichard, P. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEOTIDES. IV. ISOLATION AND CHARACTERIZATION OF THIOREDOXIN, THE HYDROGEN DONOR FROM ESCHERICHIA COLI B. J Biol Chem 1964, 239, 3436–3444. | |
| + | </ref>. | ||
| - | + | Since 1964, functions of Trx1 different than participating in cell division were discovered, as denitrosation and transnitrosation for example. Denitrosation is the removal of NO of a protein, and Trx1 does it by being temporarily S-nitrosataded on Cys32. Afterwards, Cys35 attacks Cys32 and forms a disulfide bond, releasing HNO/NO to the medium. On the other hand, transnitrosation is the the nitrosation of other proteins, that the Trx1 of some species can do. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Although Trx1 from a great amount of organisms has only the catalytic site cysteines, the human form of Trx1 also has other <scene name='91/911850/Structural_cysteines/3'>3 structural cysteines</scene>, Cys 62, Cys 69 and Cys 73, which can act as regulators of the protein (<scene name='91/911850/Trx_yeast_-_62-69-73/2'>Cys62, Cys 69 and Cys73 of Trx1 from Saccharomices cerevisiae</scene>). <scene name='91/911850/Snocys69/2'>S-nitrosation of Trx on Cys69</scene> enhances its antiapoptotic function in some cases, although its not necessary for it.<ref>Tao, L.; Gao, E.; Bryan, N. S.; Qu, Y.; Liu, H.-R.; Hu, A.; Christopher, T. A.; Lopez, B. L.; Yodoi, J.; Koch, W. J.; Feelisch, M.; Ma, X. L. Cardioprotective Effects of Thioredoxin in Myocardial Ischemia and the Reperfusion Role of S-Nitrosation. Proc Natl Acad Sci U S A 2004, 101 (31), 11471–11476. https://doi.org/10.1073/pnas.0402941101.</ref>. Cys 73 has more than one function. Firstly, it is through this residue that Trx1 transnitrosate other proteins, the Trx of not all organisms are capable of doing transnitrosation. Another function is to make Trx1 a sensor of the redox state of the cell. When the cell is in a strong oxidizing state, Trx1 forms an homodimer connected by a <scene name='91/911850/Dimer/1'>disulfide bond between the Cys73 residue of each monomer</scene>. Since Cys73 is spacially close to the active site, the formation of a dimer prevents Trx1 from interacting with other proteins and reducing them (<font color='black'><b>black</b></font> = residues Cys73, <font color='magenta'><b>pink</b></font> = active site from monomer A, <font color='orange'><b>orange</b></font> = active site from monomer B). | |
| - | == Disease == | ||
| - | = | + | <scene name='91/911850/Dipolo_positivo_no_sitio_ativo/1'>dipolo positivo no sitio ativo |
| + | </scene> | ||
| - | == | + | 1F6M ==> Thioredoxin and Thioredoxin Reductase |
| - | == | + | 2O8V ==> PAPS reductase in a covalent complex with thioredoxin C35A |
| + | 4LL4 ==> The structure of the TRX and TXNIP complex <scene name='91/911850/Trx-txnip/1'>image</scene> | ||
| - | + | 4PUF ==> Complex between the Salmonella T3SS effector SlrP and its human target thioredoxin-1 | |
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
Introduction
| |||||||||||