This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
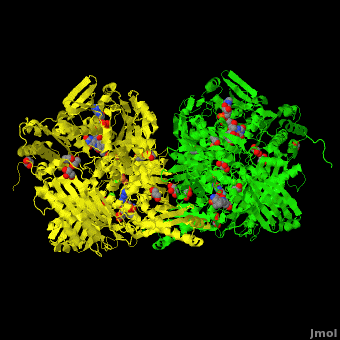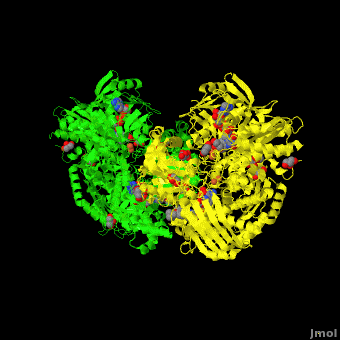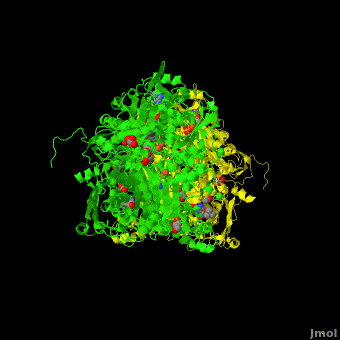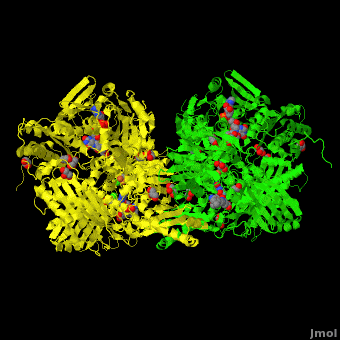
Xanthine dehydrogenase
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 18: | Line 18: | ||
substrate inhibition and in the case of 3-phenyl propionaldehyde a suicide substrate behavior. Hydroxyl-substituted aromatic aldehydes present none of these behaviors but the kinetic parameters are largely affected by the position of the OH group. High-resolution crystallographic structures obtained from single crystals of <scene name='59/599353/Cv/4'>active-DgAOR soaked with benzaldehyde</scene> showed that the side chains of Phe425 and Tyr535 are important for the stabilization of the substrate in the active site. Atoms color code: <span style="color:teal;background-color:black;font-weight:bold;">Mo in light teal</span>, <span style="color:yellow;background-color:black;font-weight:bold;">S in yellow</span>, <font color='red'><b>O in red</b></font>, <span style="color:cyan;background-color:black;font-weight:bold;">C in cyan</span> (or gray). The benzaldehyde molecules B1 and B2 were modeled in two alternative conformations (<span style="color:orange;background-color:black;font-weight:bold;">orange</span>/<span style="color:lime;background-color:black;font-weight:bold;">green</span> and <font color='navy'><b>navy</b></font>/<font color='violet'><b>violet</b></font>, | substrate inhibition and in the case of 3-phenyl propionaldehyde a suicide substrate behavior. Hydroxyl-substituted aromatic aldehydes present none of these behaviors but the kinetic parameters are largely affected by the position of the OH group. High-resolution crystallographic structures obtained from single crystals of <scene name='59/599353/Cv/4'>active-DgAOR soaked with benzaldehyde</scene> showed that the side chains of Phe425 and Tyr535 are important for the stabilization of the substrate in the active site. Atoms color code: <span style="color:teal;background-color:black;font-weight:bold;">Mo in light teal</span>, <span style="color:yellow;background-color:black;font-weight:bold;">S in yellow</span>, <font color='red'><b>O in red</b></font>, <span style="color:cyan;background-color:black;font-weight:bold;">C in cyan</span> (or gray). The benzaldehyde molecules B1 and B2 were modeled in two alternative conformations (<span style="color:orange;background-color:black;font-weight:bold;">orange</span>/<span style="color:lime;background-color:black;font-weight:bold;">green</span> and <font color='navy'><b>navy</b></font>/<font color='violet'><b>violet</b></font>, | ||
respectively) On the other hand, the X-ray data of <scene name='59/599353/Cv/5'>DgAOR soaked with trans-cinnamaldehyde</scene> showed a cinnamic acid molecule in the substrate channel. <span style="color:orange;background-color:black;font-weight:bold;">In orange an alternate conformation of the side chain of Phe425 and a molecule of trans-cinnamaldehyde</span>. The X-ray data of <scene name='59/599353/Cv/7'>DgAOR soaked with 3-phenyl propionaldehyde</scene> showed clearly how high substrate concentrations inactivate the enzyme by binding covalently at the surface of the enzyme and blocking the substrate channel. The different reactivity of DgAOR versus aldehyde oxidase and XO towards aromatic aldehydes and N-heterocyclic compounds is explained on the basis of the present kinetic and structural data. | respectively) On the other hand, the X-ray data of <scene name='59/599353/Cv/5'>DgAOR soaked with trans-cinnamaldehyde</scene> showed a cinnamic acid molecule in the substrate channel. <span style="color:orange;background-color:black;font-weight:bold;">In orange an alternate conformation of the side chain of Phe425 and a molecule of trans-cinnamaldehyde</span>. The X-ray data of <scene name='59/599353/Cv/7'>DgAOR soaked with 3-phenyl propionaldehyde</scene> showed clearly how high substrate concentrations inactivate the enzyme by binding covalently at the surface of the enzyme and blocking the substrate channel. The different reactivity of DgAOR versus aldehyde oxidase and XO towards aromatic aldehydes and N-heterocyclic compounds is explained on the basis of the present kinetic and structural data. | ||
| - | </StructureSection> | ||
==3D structures of xanthine dehydrogenase== | ==3D structures of xanthine dehydrogenase== | ||
| - | + | [[Xanthine dehydrogenase 3D structures]] | |
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== References == | == References == | ||
Current revision
| |||||||||||
References
- ↑ B-Rao C, Kulkarni-Almeida A, Katkar KV, Khanna S, Ghosh U, Keche A, Shah P, Srivastava A, Korde V, Nemmani KV, Deshmukh NJ, Dixit A, Brahma MK, Bahirat U, Doshi L, Sharma R, Sivaramakrishnan H. Identification of novel isocytosine derivatives as xanthine oxidase inhibitors from a set of virtual screening hits. Bioorg Med Chem. 2012 May 1;20(9):2930-9. Epub 2012 Mar 14. PMID:22483591 doi:10.1016/j.bmc.2012.03.019
- ↑ Pauff JM, Cao H, Hille R. Substrate Orientation and Catalysis at the Molybdenum Site in Xanthine Oxidase: CRYSTAL STRUCTURES IN COMPLEX WITH XANTHINE AND LUMAZINE. J Biol Chem. 2009 Mar 27;284(13):8760-7. Epub 2008 Dec 24. PMID:19109252 doi:10.1074/jbc.M804517200
- ↑ Dietzel U, Kuper J, Doebbler JA, Schulte A, Truglio JJ, Leimkuhler S, Kisker C. Mechanism of Substrate and Inhibitor Binding of Rhodobacter capsulatus Xanthine Dehydrogenase. J Biol Chem. 2009 Mar 27;284(13):8768-76. Epub 2008 Dec 24. PMID:19109249 doi:http://dx.doi.org/10.1074/jbc.M808114200
- ↑ Fukunari A, Okamoto K, Nishino T, Eger BT, Pai EF, Kamezawa M, Yamada I, Kato N. Y-700 [1-[3-Cyano-4-(2,2-dimethylpropoxy)phenyl]-1H-pyrazole-4-carboxylic acid]: a potent xanthine oxidoreductase inhibitor with hepatic excretion. J Pharmacol Exp Ther. 2004 Nov;311(2):519-28. Epub 2004 Jun 9. PMID:15190124 doi:10.1124/jpet.104.070433
- ↑ 5.0 5.1 Okamoto K, Eger BT, Nishino T, Kondo S, Pai EF, Nishino T. An extremely potent inhibitor of xanthine oxidoreductase. Crystal structure of the enzyme-inhibitor complex and mechanism of inhibition. J Biol Chem. 2003 Jan 17;278(3):1848-55. Epub 2002 Nov 5. PMID:12421831 doi:10.1074/jbc.M208307200
- ↑ Truglio JJ, Theis K, Leimkuhler S, Rappa R, Rajagopalan KV, Kisker C. Crystal structures of the active and alloxanthine-inhibited forms of xanthine dehydrogenase from Rhodobacter capsulatus. Structure. 2002 Jan;10(1):115-25. PMID:11796116
- ↑ 7.0 7.1 Okamoto K, Matsumoto K, Hille R, Eger BT, Pai EF, Nishino T. The crystal structure of xanthine oxidoreductase during catalysis: implications for reaction mechanism and enzyme inhibition. Proc Natl Acad Sci U S A. 2004 May 25;101(21):7931-6. Epub 2004 May 17. PMID:15148401 doi:10.1073/pnas.0400973101
- ↑ Correia HD, Marangon J, Brondino CD, Moura JJ, Romao MJ, Gonzalez PJ, Santos-Silva T. Aromatic aldehydes at the active site of aldehyde oxidoreductase from Desulfovibrio gigas: reactivity and molecular details of the enzyme-substrate and enzyme-product interaction. J Biol Inorg Chem. 2014 Sep 27. PMID:25261288 doi:http://dx.doi.org/10.1007/s00775-014-1196-4