User:Arthur Migliatti/Sandbox1
From Proteopedia
< User:Arthur Migliatti(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 12: | Line 12: | ||
== Structure == | == Structure == | ||
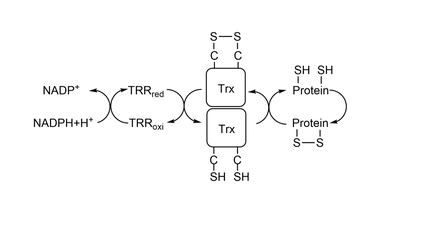
| - | Trx1 is a monomeric protein and weights around 12kDa. It is formed by one five-stranded beta sheet involved by 4 alpha helix, shown <scene name='91/911850/Secondary_structure/1'>here</scene>. The active site is located on a lump between betra strand 2, where Cys 35 is located, and alpha helix 2, where Cys32 is located. By being in the end of an alpha helix, <scene name='91/911850/Secondary_structure_cys32/1'>Cys32</scene> has a lower pKa, making it possible to reduce disulfide bonds | + | Trx1 is a monomeric protein and weights around 12kDa. It is formed by one five-stranded beta sheet involved by 4 alpha helix, shown <scene name='91/911850/Secondary_structure/1'>here</scene>. The active site is located on a lump between betra strand 2, where Cys 35 is located, and alpha helix 2, where Cys32 is located. By being in the end of an alpha helix, <scene name='91/911850/Secondary_structure_cys32/1'>Cys32</scene> has a lower pKa, making it possible to reduce disulfide bonds<ref>Holmgren, A. Thioredoxin Structure and Mechanism: Conformational Changes on Oxidation of the Active-Site Sulfhydryls to a Disulfide. Structure 1995, 3 (3), 239–243. https://doi.org/10.1016/S0969-2126(01)00153-8.</ref>. As it is shown in this <scene name='91/911850/Dipolo_positivo_no_sitio_ativo/1'>image</scene>, the nitrogens point to the active site, while the oxigens point to the other direction. This creates a positive dipole, which reduces the pKa of the active site. |
| - | </ref> | + | |
The structure of the protein doesn't change when it goes from reduced to oxidized, but the sulfur of the cysteines in the active site come closer, although both cysteines stay in the same distance(<scene name='91/911850/Trx-oxi-dislig-s-s_-_ca-ca/2'>oxidized</scene> x <scene name='91/911850/Trx-cys-red-dislig-s-ca/3'>reduced</scene>). As it's possible to see in the next images, the dihedral angle made by N - Calpha - C - S changes in both cysteines. In the <scene name='91/911850/Trx-cys-red-ang-s-ca/1'>reduced state</scene>, the angles are in such way that the sulfur atoms are far apart. When it is in the <scene name='91/911850/Trx-oxi-dislig-s-s_-_ca-ca/3'>oxidized state</scene>, the sulfur atoms come closer together and bond. | The structure of the protein doesn't change when it goes from reduced to oxidized, but the sulfur of the cysteines in the active site come closer, although both cysteines stay in the same distance(<scene name='91/911850/Trx-oxi-dislig-s-s_-_ca-ca/2'>oxidized</scene> x <scene name='91/911850/Trx-cys-red-dislig-s-ca/3'>reduced</scene>). As it's possible to see in the next images, the dihedral angle made by N - Calpha - C - S changes in both cysteines. In the <scene name='91/911850/Trx-cys-red-ang-s-ca/1'>reduced state</scene>, the angles are in such way that the sulfur atoms are far apart. When it is in the <scene name='91/911850/Trx-oxi-dislig-s-s_-_ca-ca/3'>oxidized state</scene>, the sulfur atoms come closer together and bond. | ||
| Line 34: | Line 33: | ||
2O8V ==> PAPS reductase in a covalent complex with thioredoxin C35A | 2O8V ==> PAPS reductase in a covalent complex with thioredoxin C35A | ||
| - | 4LL4 ==> The structure of the TRX and TXNIP complex | + | 4LL4 ==> The structure of the TRX and TXNIP complex <scene name='91/911850/Trx-txnip/1'>image</scene> |
4PUF ==> Complex between the Salmonella T3SS effector SlrP and its human target thioredoxin-1 | 4PUF ==> Complex between the Salmonella T3SS effector SlrP and its human target thioredoxin-1 | ||
Current revision
Introduction
| |||||||||||