This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox reserved 1752
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
<structuresection load='2IS4' size='340' side='right'caption='[[2IS4]], [[Resolution|resolution]] 2.70Å' scene=''> | <structuresection load='2IS4' size='340' side='right'caption='[[2IS4]], [[Resolution|resolution]] 2.70Å' scene=''> | ||
| - | == UvrD == | ||
| - | |||
| + | Copy From here down | ||
| + | == UvrD == | ||
<scene name='92/925553/Uvrd/1'>UvrD</scene>, also known as Helicase II, is one of many components responsible in repairing DNA damage. Helicases use energy from ATP to unwind double helices in metabolic pathways using nucleic acids. ATP molecules are typically used to store energy shared between phosphate groups that gets released when breaking bonds to drive catabolic reactions. | <scene name='92/925553/Uvrd/1'>UvrD</scene>, also known as Helicase II, is one of many components responsible in repairing DNA damage. Helicases use energy from ATP to unwind double helices in metabolic pathways using nucleic acids. ATP molecules are typically used to store energy shared between phosphate groups that gets released when breaking bonds to drive catabolic reactions. | ||
| - | Helicases were found in the 1970’s to be DNA-dependent ATPases, meaning that they use ATP hydrolysis to complete its interactions with the different types of nucleic acids it comes into contact with. Helicase II, also called UvrD is the founding member of SF1, one group of six superfamiliies used to identify helicases. SF1 and SF2 members share seven conserved sequence motifs that are involved in ATP Binding <ref name="ATP Binding">PMID:17190599</ref>. UvrD is important in replication, recombination, and repair from ultraviolet damage and mismatched base pairs. Nucleotide excision repair in a normal cell is supposed to correct pyrimidine dimers and other DNA lesions when bases are displaced from their normal positions. UvrD pairs up with the UvrABC endonuclease system, which works to displace the DNA. This is then repaired by PolI and DNA ligase | + | Helicases were found in the 1970’s to be DNA-dependent ATPases, meaning that they use ATP hydrolysis to complete its interactions with the different types of nucleic acids it comes into contact with. Helicase II, also called UvrD is the founding member of SF1, one group of six superfamiliies used to identify helicases. SF1 and SF2 members share seven conserved sequence motifs that are involved in ATP Binding <ref name="ATP Binding">PMID:17190599</ref>. UvrD is important in replication, recombination, and repair from ultraviolet damage and mismatched base pairs. Nucleotide excision repair in a normal cell is supposed to correct pyrimidine dimers and other DNA lesions when bases are displaced from their normal positions. UvrD pairs up with the UvrABC endonuclease system, which works to displace the DNA. This is then repaired by PolI and DNA ligase <ref>Voet, D., Voet, J., & Pratt, C. (2015). Fundamentals of Biochemistry: Life at the Molecular Level (4th ed.). Wiley</ref>. |
| - | + | ||
== UvrD Motifs == | == UvrD Motifs == | ||
There are<scene name='92/925553/Uvrd_labeled_motifs_complete/3'> 16 binding motifs</scene> for UvrD, which are conserved in other homologous structures. The homologous structures mentioned are Helicase 2 homologs, which appear in different species. These conserved motifs are important to maintain the function of UvrD. There are 4 domains that these motifs fit into (not shown). The domains are 1A, 1B, 2A, and 2B. Motifs I, Ia, II-VI are involved in ATP binding. Motifs Ia, III, and V are involved in ssDNA binding. Motif IV is reported to be unique in SF1. They found in their paper, seven new sequence motifs conserved among UvrD homologs. They are Ib, Ic, Id, IVb, IVc, Va, and VIa. These conserved residues are involved in DNA binding or domain 1B and 2B interactions <ref name="ATP_Binding" />. | There are<scene name='92/925553/Uvrd_labeled_motifs_complete/3'> 16 binding motifs</scene> for UvrD, which are conserved in other homologous structures. The homologous structures mentioned are Helicase 2 homologs, which appear in different species. These conserved motifs are important to maintain the function of UvrD. There are 4 domains that these motifs fit into (not shown). The domains are 1A, 1B, 2A, and 2B. Motifs I, Ia, II-VI are involved in ATP binding. Motifs Ia, III, and V are involved in ssDNA binding. Motif IV is reported to be unique in SF1. They found in their paper, seven new sequence motifs conserved among UvrD homologs. They are Ib, Ic, Id, IVb, IVc, Va, and VIa. These conserved residues are involved in DNA binding or domain 1B and 2B interactions <ref name="ATP_Binding" />. | ||
== Separation Pin == | == Separation Pin == | ||
| - | The "<scene name='92/925553/Pin_complex/ | + | The "<scene name='92/925553/Pin_complex/2'>separation pin</scene>" is a part of the 2B domain and is responsible for unwinding the DNA. This uses a 2 step power stroke, one stroke when ATP is bound and another stroke when ADP and P<sub>i</sub> are released. The GIG motif and separation pin work together to unwind the DNA and move it out of the way so UvrD can unwind more DNA. The separation pin also prevents ssDNA once unwound from moving backwards and from reannealing. The proposed method is called the wrench-and-inchworm method, which is when the enzyme binds DNA and attaches at different points and then moves 1 nucleotide per ATP molecule.After an ATP molecule is released, UvrD is then ready to proceed forward to the next nucleotide <ref name="ATP_Binding" />. |
== UvrD Binding Site for ATP analog (AMPPNP) == | == UvrD Binding Site for ATP analog (AMPPNP) == | ||
When determining the structure of UvrD, an ATP analog was used. They used an <scene name='92/925553/Atp_analog/3'>ATP analog</scene> so that the last phosphate can't be cleaved. Using the unhydrolyzable analog is beneficial in locking in the structure to observe.The green ion shown in the ATP analog scene is a Mg<sup>2+</sup> ion, which is essential for ATP hydrolysis and interacts with the β and γ phosphates. The magnesium ion is surrounded by essential residues that when altered, have been shown to have reduced ATPase activity <ref name="ATP_Binding" />. | When determining the structure of UvrD, an ATP analog was used. They used an <scene name='92/925553/Atp_analog/3'>ATP analog</scene> so that the last phosphate can't be cleaved. Using the unhydrolyzable analog is beneficial in locking in the structure to observe.The green ion shown in the ATP analog scene is a Mg<sup>2+</sup> ion, which is essential for ATP hydrolysis and interacts with the β and γ phosphates. The magnesium ion is surrounded by essential residues that when altered, have been shown to have reduced ATPase activity <ref name="ATP_Binding" />. | ||
== UvrD Binding Site for ATP analog (ADP•MgF<sub>3</sub>) == | == UvrD Binding Site for ATP analog (ADP•MgF<sub>3</sub>) == | ||
To capture the UvrD-DNA-ADP complex, a new crystal structure used ADP•MgF<sub>3</sub> after NaF was added to help improve crystal growth. This structure is believed to be a more authentic transition state analog, which differs from the AMPPNP analog slightly. The <scene name='92/925553/Adp_analog_complete/1'>ADP analog</scene> has a <scene name='92/925553/Adp_e566_and_gol/2'>GOL region</scene>, which is a glycerol molecule, which has hydrogen bonding similar to interactions that E566 has to a 3' OH of the ribose. The DNA isn't actually bound in the crystal structure, but can be used to visualize what hydrogen bonding might look like when connected to the backbone in DNA. <scene name='92/925553/Adp_e566_and_gol_hbonding_comp/2'>This glycerol molecule hydrogen bonds with E566</scene>, which typically would bind to the 3' OH of the ribose of DNA. Another residue, R37 (Not Shown), binds to the 2' OH of ribose, which has weaker hydrogen bonding. This is a structural component that allows UvrD to bind both ATP and dATP<ref name="ATP_Binding" />. | To capture the UvrD-DNA-ADP complex, a new crystal structure used ADP•MgF<sub>3</sub> after NaF was added to help improve crystal growth. This structure is believed to be a more authentic transition state analog, which differs from the AMPPNP analog slightly. The <scene name='92/925553/Adp_analog_complete/1'>ADP analog</scene> has a <scene name='92/925553/Adp_e566_and_gol/2'>GOL region</scene>, which is a glycerol molecule, which has hydrogen bonding similar to interactions that E566 has to a 3' OH of the ribose. The DNA isn't actually bound in the crystal structure, but can be used to visualize what hydrogen bonding might look like when connected to the backbone in DNA. <scene name='92/925553/Adp_e566_and_gol_hbonding_comp/2'>This glycerol molecule hydrogen bonds with E566</scene>, which typically would bind to the 3' OH of the ribose of DNA. Another residue, R37 (Not Shown), binds to the 2' OH of ribose, which has weaker hydrogen bonding. This is a structural component that allows UvrD to bind both ATP and dATP<ref name="ATP_Binding" />. | ||
| + | <scene name='92/925553/Uvrd/2'>UvrD</scene> | ||
| + | <scene name='92/925553/Atp_analog/6'>ATP analog</scene> | ||
| + | Stop here | ||
| + | ==Cystic fibrosis transmembrane conductance regulator (CFTR)== | ||
| + | <StructureSection load='5UAK' size='340' side='right' caption='Cystic Fibrosis Transmembrane Conductance regulator' scene=''> | ||
| + | The '''CFTR''' is a chloride channel, and is regulated by PKA phosphorylation, cAMP levels, and ATP/ADP ratios. Mutations in the CFTR cause the disease cystic fibrosis. | ||
| + | |||
| + | CFTR is a mostly <scene name='78/785332/Secondary_structure/1'>alpha helical</scene> protein. The membrane spanning segments can be clearly seen with coloring by <scene name='78/785332/Hydrophobicity/1'>hydrophobicity</scene>, which shows hydrophobic residues in gray and hydrophilic residues in purple. | ||
| + | |||
| + | The extracellular end of the channel has several <scene name='78/785332/Ec_cl_selection/1'>positively charged</scene> residues that are important for recruiting chloride ions to the channel. A number of <scene name='78/785332/Plus_channel/1'>positively charged</scene> residues line the channel. In the unphosphorylated state (as this structure is), a <scene name='78/785332/Regulatory_domain/2'>regulatory domain</scene> blocks the activity of the channel (the connecting segments are not visible in the structure). It contains several negatively charged residues; when the protein is phosphorylated, this segment is repelled, causing a structural change. <ref>PMID:28340353</ref> | ||
| + | |||
| + | CFTR contains two <scene name='78/785332/Nbd/2'>nucleotide binding domains</scene> (NBD's), which both contain <scene name='78/785332/Walker_motifs/2'>Walker motifs</scene>, flexible loops that bind phosphate groups tightly and are highly conserved among ATP-binding proteins. | ||
| + | |||
| + | ==Mutations in Cystic Fibrosis== | ||
| + | |||
| + | Cystic fibrosis is characterized by decreased chloride transport, which causes mucus to be thicker and stickier. This leads to a variety of problems, including decreased lung capacity, decreased pancreatic enzyme release into the small intestine, increased rates of lung infections, and infertility.<ref>https://ghr.nlm.nih.gov/condition/cystic-fibrosis</ref> There are a wide assortment of mutations that cause cystic fibrosis, with differing symptom severity. The deletion of <scene name='78/785332/F508/1'>F508</scene> causes the protein to not be properly synthesized, and no expression is seen on the cell surface. Other mutations are found in the NBD's; some of these mutations such as S1255P alter the responsiveness to MgATP, while others such as G551S, G1244E, and G1239D decrease the frequency of channel opening. | ||
| + | |||
| + | Some of the mutations that lead to cystic fibrosis are due to folding errors. There are <scene name='78/785332/Numbered_bundles/2'>12 transmembrane sequences</scene> in CFTR; they are not sequential in their packing. The presence of <scene name='78/785332/Numbered_bundles_pos_res/1'>hydrophilic, positively charged amino acids</scene> in these transmembrane sequences (shown in red) lead to a folding problem: how do you stabilize them until they can be protected by hydrophobic residues and are no longer exposed to the hydrophobic membrane? | ||
| + | |||
| + | ==3D Printed Physical Model of the CFTR protein== | ||
| + | |||
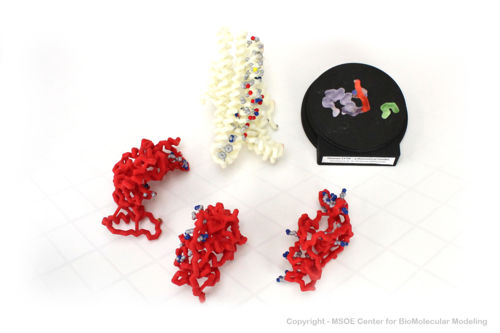
| + | Shown below are 3D printed physical models of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) protein. The backbone model on the left is colored by region, with the transmembrane domain white and the atp-binding and regulatory domains colored red. The backbone model on the right is colored by regional repeat, with the first repeat blue, the second repeat green and the regulatory domain colored red. The models have been designed with embedded magnets to disassemble into the key regions of the structure. | ||
| + | |||
| + | [[Image:cftr1_centerForBioMolecularModeling.jpg|500px]] | ||
| + | [[Image:cftr2_centerForBioMolecularModeling.jpg|500px]] | ||
| + | |||
| + | ====The MSOE Center for BioMolecular Modeling==== | ||
| + | |||
| + | [[Image:CbmUniversityLogo.jpg | left | 150px]] | ||
| + | |||
| + | The [http://cbm.msoe.edu MSOE Center for BioMolecular Modeling] uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our [http://cbm.msoe.edu/educationalmedia/modelgallery/ Model Gallery]. | ||
| + | |||
| + | |||
| + | |||
| + | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references /> | <references /> | ||
| - | Voet, Voet, & Pratt. (n.d.). Fundamentals biochemistry 4e : Free download, borrow, and streaming. Internet Archive. Retrieved October 11, 2022, from https://archive.org/details/FundamentalsBiochemistry4e | ||
Current revision
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006 Dec 29;127(7):1349-60. PMID:17190599 doi:http://dx.doi.org/10.1016/j.cell.2006.10.049
- ↑ Voet, D., Voet, J., & Pratt, C. (2015). Fundamentals of Biochemistry: Life at the Molecular Level (4th ed.). Wiley
- ↑ Liu F, Zhang Z, Csanady L, Gadsby DC, Chen J. Molecular Structure of the Human CFTR Ion Channel. Cell. 2017 Mar 23;169(1):85-95.e8. doi: 10.1016/j.cell.2017.02.024. PMID:28340353 doi:http://dx.doi.org/10.1016/j.cell.2017.02.024
- ↑ https://ghr.nlm.nih.gov/condition/cystic-fibrosis