NKX2.5 Homeodomain
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 5: | Line 5: | ||
[[Image:Map.png |thumb|right|upright=3.5|''Organization of the biological protein NKX2.5. The structures analyzed only represent the homeodomain region.'']] | [[Image:Map.png |thumb|right|upright=3.5|''Organization of the biological protein NKX2.5. The structures analyzed only represent the homeodomain region.'']] | ||
| - | The transcription factor, NKX2.5 is one of many proteins classified as a homeodomain, and functions to regulate structural development in eukaryotes. These proteins share a characteristic evolutionarily conserved fold containing <scene name='91/911264/Three_alpha_helices/2'>three alpha-helices</scene>. <ref name="WJ"> PMID: 7979246 </ref>. DNA-binding is mediated through the insertion of the C-terminal side <scene name='91/911264/Major_groove_interaction/4'>alpha-helix</scene> (alpha-3) into the major groove, allowing for base-reside interactions. This allows homeodomains to locate and bind specific DNA sequences, leading to transcriptional activation or repression <ref> PMID: 26464018 </ref>. The homeodomain of NKX2.5 is flanked by both a N and C-terminal regulatory domain. This puts the biological protein at 324 residues with the homeodomain consisting of residues 138-197 <ref> PMID: 22849347 </ref> Research into the structure and function of NKX2.5 has mainly been focused on the DNA-binding homeodomain, as mutations in this region have been linked to specific diseases <ref name="Schott"> PMID: 9651244</ref>. This page will focus on the specific interactions of the homeodomain of NKX2.5 with DNA, and how these interactions relate to one of the transcription factor's primary function - heart development. | + | The transcription factor, '''NKX2.5''' is one of many proteins classified as a homeodomain, and functions to regulate structural development in eukaryotes. These proteins share a characteristic evolutionarily conserved fold containing <scene name='91/911264/Three_alpha_helices/2'>three alpha-helices</scene>. <ref name="WJ"> PMID: 7979246 </ref>. DNA-binding is mediated through the insertion of the C-terminal side <scene name='91/911264/Major_groove_interaction/4'>alpha-helix</scene> (alpha-3) into the major groove, allowing for base-reside interactions. This allows homeodomains to locate and bind specific DNA sequences, leading to transcriptional activation or repression <ref> PMID: 26464018 </ref>. The homeodomain of NKX2.5 is flanked by both a N and C-terminal regulatory domain. This puts the biological protein at 324 residues with the homeodomain consisting of residues 138-197 <ref> PMID: 22849347 </ref> Research into the structure and function of NKX2.5 has mainly been focused on the DNA-binding homeodomain, as mutations in this region have been linked to specific diseases <ref name="Schott"> PMID: 9651244</ref>. This page will focus on the specific interactions of the homeodomain of NKX2.5 with DNA, and how these interactions relate to one of the transcription factor's primary function - heart development. |
| - | + | = Clinical Relevance = | |
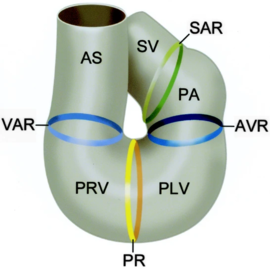
[[Image:CardiacLooping.png|thumb|right|upright=1.5|''The sinus venosus (SV), the sinoatrial ring (SAR), the primitive atrium (PA), atrioventricular ring (AVR), primitive left ventricle (PLV), primary ring (PR), primitive right ventricle (PRV), ventriculoarterial ring (VAR), and the aortic sac (AS) <ref name="Groot" />.'']] | [[Image:CardiacLooping.png|thumb|right|upright=1.5|''The sinus venosus (SV), the sinoatrial ring (SAR), the primitive atrium (PA), atrioventricular ring (AVR), primitive left ventricle (PLV), primary ring (PR), primitive right ventricle (PRV), ventriculoarterial ring (VAR), and the aortic sac (AS) <ref name="Groot" />.'']] | ||
In the case of NKX2.5, the protein works in conjunction with multiple other transcription factors during cardiogenesis <ref> PMID: 17008524</ref>,<ref> PMID: 15925411 </ref>. Recently, research has been focused on NKX2.5 as mutations in the DNA binding residues, and structural support residues of the protein have been linked to congenital heart disease. Specifically, NKX2.5 mutations have been linked to etiologies involving both atrial and septal defects, deficient atrioventricular node conduction, and more complex mutations such as Tetralogy of Fallot and Hypoplastic Left Heart Syndrome <ref> PMID: 11889119</ref>,<ref name="Schott" />,<ref> PMID: 14607454 </ref>. These phenotypes are speculated to arise as a result of decreased DNA-binding of NKX2.5 during cardiogenesis. Thi <ref> Carlson, B. M. (1994). Human embryology and developmental biology. St. Louis: Mosby. </ref>. Specifically, issues take root during cardiac loop formation, which takes place approximately 24 days after embryogenesis. At this time, the single vessel present in the embryo loops back upon itself, allowing the vessel to come in contact with itself. This leads to the formation of a four-chambered, primitive heart with the great vessels (shown in the figure from Gittenberger-de Groot ''et. al'')<ref name="Groot"> PMID: 15611355 </ref>. Throughout this point in development, NKX2.5 along with other transcription factors are essential in facilitating initial loop formation. Therefore, mutations that affect NKX2.5 function have been tied to the defects mentioned above <ref> PMID: 10021345 </ref>. | In the case of NKX2.5, the protein works in conjunction with multiple other transcription factors during cardiogenesis <ref> PMID: 17008524</ref>,<ref> PMID: 15925411 </ref>. Recently, research has been focused on NKX2.5 as mutations in the DNA binding residues, and structural support residues of the protein have been linked to congenital heart disease. Specifically, NKX2.5 mutations have been linked to etiologies involving both atrial and septal defects, deficient atrioventricular node conduction, and more complex mutations such as Tetralogy of Fallot and Hypoplastic Left Heart Syndrome <ref> PMID: 11889119</ref>,<ref name="Schott" />,<ref> PMID: 14607454 </ref>. These phenotypes are speculated to arise as a result of decreased DNA-binding of NKX2.5 during cardiogenesis. Thi <ref> Carlson, B. M. (1994). Human embryology and developmental biology. St. Louis: Mosby. </ref>. Specifically, issues take root during cardiac loop formation, which takes place approximately 24 days after embryogenesis. At this time, the single vessel present in the embryo loops back upon itself, allowing the vessel to come in contact with itself. This leads to the formation of a four-chambered, primitive heart with the great vessels (shown in the figure from Gittenberger-de Groot ''et. al'')<ref name="Groot"> PMID: 15611355 </ref>. Throughout this point in development, NKX2.5 along with other transcription factors are essential in facilitating initial loop formation. Therefore, mutations that affect NKX2.5 function have been tied to the defects mentioned above <ref> PMID: 10021345 </ref>. | ||
| Line 14: | Line 14: | ||
= Structural highlights = | = Structural highlights = | ||
| - | + | == Protein / DNA Interactions == | |
DNA-binding is mediated by residues of <scene name='91/911264/Major_groove_interaction/4'>alpha-helix three</scene> probing into the major groove. In the structure presented, NKX2.5 is binding to a known target: the ANF-242 sequence downstream of the atrial natriuretic factor peptide promoter <ref name="Schott" />. The ANF-242 site contains two NK family binding motifs, <scene name='91/911264/Nk_motif/1'>AAGTG</scene>, separated by a five nucelotide gap <ref> PMID: 21930795 </ref>. NKX2.5 interacts to this motif with residues positioned on alpha helix 3. Thre residues, <scene name='91/911264/Dna-backbone_interactions/1'>Gln 187, Arg 190 and Lys 194</scene> interact with DNA backbone at the NK binding motif. Interactions to bases in the major groove is mediated by two residues: <scene name='91/911264/Major_groove_interactions/2'>Asn 188 and Tyr 191</scene>. In addition to these two major grove interactions, there are three additional reidues that interact with bases in the NK motif. These bases are <scene name='91/911264/Other_residues/1'>Arg 142, Ile 184 and Gln 50</scene>. These residues are of particular importance as mutations in this region has been directly linked to pathologies discussed above <ref name="Schott" />,<ref> PMID: 26926761 </ref>. | DNA-binding is mediated by residues of <scene name='91/911264/Major_groove_interaction/4'>alpha-helix three</scene> probing into the major groove. In the structure presented, NKX2.5 is binding to a known target: the ANF-242 sequence downstream of the atrial natriuretic factor peptide promoter <ref name="Schott" />. The ANF-242 site contains two NK family binding motifs, <scene name='91/911264/Nk_motif/1'>AAGTG</scene>, separated by a five nucelotide gap <ref> PMID: 21930795 </ref>. NKX2.5 interacts to this motif with residues positioned on alpha helix 3. Thre residues, <scene name='91/911264/Dna-backbone_interactions/1'>Gln 187, Arg 190 and Lys 194</scene> interact with DNA backbone at the NK binding motif. Interactions to bases in the major groove is mediated by two residues: <scene name='91/911264/Major_groove_interactions/2'>Asn 188 and Tyr 191</scene>. In addition to these two major grove interactions, there are three additional reidues that interact with bases in the NK motif. These bases are <scene name='91/911264/Other_residues/1'>Arg 142, Ile 184 and Gln 50</scene>. These residues are of particular importance as mutations in this region has been directly linked to pathologies discussed above <ref name="Schott" />,<ref> PMID: 26926761 </ref>. | ||
| - | + | == Protein / Protein Interactions== | |
The global structure of NKX2.5 is maintained by both hydrophobic and hydrophilic interactions between the three alpha-helices. This is clear when observing the structure of NKX2.5 where <scene name='91/911264/Hydrophilic_and_phobic/1'>coloring is based on hydrophobicity</scene>. Besides these tertiary interactions present in NKX2.5, the protein is known to associate with other transcription factors and regulatory proteins. For example, GATA factors, and the Hand1 transcription factor (both of which are important in cardiogenesis) are known to interact with NKX2.5 <ref> PMID: 9312027 </ref>,<ref> PMID: 21519287 </ref>. Unfortunately, structures are not available for these complexes. Structures have been elucidated for the interaction of <scene name='91/911264/Tbx5/1'>NKX2.5 with the transcription factor TBX5</scene> (T-box5)<ref> PMID: 26926761 </ref>. This interaction appears is thought to be mediated <scene name='91/911264/Tbx5_interactions/1'>through three residues</scene>: Lys 158 of NKX2.5 along with Asp 140 and Pro 142 from TBX5. A complex formed between <scene name='91/911264/Nkx25_and_mef2/1'>NKX2.5 and MEF2</scene> (myocyte enhancer factor 2) has also been elucidated <ref> PMID: 32681840 </ref>. | The global structure of NKX2.5 is maintained by both hydrophobic and hydrophilic interactions between the three alpha-helices. This is clear when observing the structure of NKX2.5 where <scene name='91/911264/Hydrophilic_and_phobic/1'>coloring is based on hydrophobicity</scene>. Besides these tertiary interactions present in NKX2.5, the protein is known to associate with other transcription factors and regulatory proteins. For example, GATA factors, and the Hand1 transcription factor (both of which are important in cardiogenesis) are known to interact with NKX2.5 <ref> PMID: 9312027 </ref>,<ref> PMID: 21519287 </ref>. Unfortunately, structures are not available for these complexes. Structures have been elucidated for the interaction of <scene name='91/911264/Tbx5/1'>NKX2.5 with the transcription factor TBX5</scene> (T-box5)<ref> PMID: 26926761 </ref>. This interaction appears is thought to be mediated <scene name='91/911264/Tbx5_interactions/1'>through three residues</scene>: Lys 158 of NKX2.5 along with Asp 140 and Pro 142 from TBX5. A complex formed between <scene name='91/911264/Nkx25_and_mef2/1'>NKX2.5 and MEF2</scene> (myocyte enhancer factor 2) has also been elucidated <ref> PMID: 32681840 </ref>. | ||
| - | + | = Evolution = | |
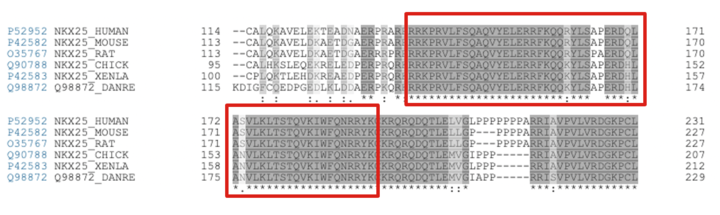
The homeodomain of NKX2.5 is highly conserved across the animal kingdom <ref name="WJ" />. This can easily be seen in a sequence alignment containing NKX2.5 isoforms across a variety of species. A selection of the Clustal Omega sequence alignment <ref> PMID: 23671338 </ref> shown below outlines the conservation of the homeodomain of NKX2.5. | The homeodomain of NKX2.5 is highly conserved across the animal kingdom <ref name="WJ" />. This can easily be seen in a sequence alignment containing NKX2.5 isoforms across a variety of species. A selection of the Clustal Omega sequence alignment <ref> PMID: 23671338 </ref> shown below outlines the conservation of the homeodomain of NKX2.5. | ||
| Line 25: | Line 25: | ||
It is important to note that the residues involved in the recognition of the NKX2.5 motif are fully conserved across all isoforms. The cross-species similarities in NKX2.5 isozymes is of special importance in relation to research. For example, ''Danio rerio'' (Zebrafish) have been used as models to study heart development during gestation <ref> PMID: 10993952 </ref>, <ref> PMID: 28615160 </ref>. This is made possible by the fact that the ''Danio rerio'' and ''Homo sapien'' NKX2.5 share sequence similarities. | It is important to note that the residues involved in the recognition of the NKX2.5 motif are fully conserved across all isoforms. The cross-species similarities in NKX2.5 isozymes is of special importance in relation to research. For example, ''Danio rerio'' (Zebrafish) have been used as models to study heart development during gestation <ref> PMID: 10993952 </ref>, <ref> PMID: 28615160 </ref>. This is made possible by the fact that the ''Danio rerio'' and ''Homo sapien'' NKX2.5 share sequence similarities. | ||
| - | == | + | =Homeobox protein Nkx-2.5 3D structures= |
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| + | |||
| + | [[3rkq]] – hNkx-2.5 + DNA – human <br /> | ||
| + | [[6wc2]], [[6wc5]] – hNkx-2.5 + myocyte-specific enhancer factor 2B + DNA <br /> | ||
| + | [[4s0h]] – hNkx-2.5 + TBX5 + DNA <br /> | ||
| + | [[5flv]] – Nkx-2.5 + TBX5 + DNA - mouse<br /> | ||
| + | [[2l9r]] – hNkx-3.1 - NMR <br /> | ||
| + | |||
| + | = References = | ||
<references/> | <references/> | ||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||