User:Jackson Langford/Sandbox 1
From Proteopedia
< User:Jackson Langford(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
=''HUMAN'' THYROTROPIN RECEPTOR= | =''HUMAN'' THYROTROPIN RECEPTOR= | ||
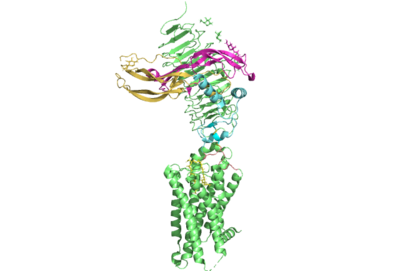
| - | <StructureSection load='7t9m' size='350' frame='true' side='right' caption='Thyrotropin Receptor 7T9M' scene='95/954073/The_residues_of_p10_peptide/4'> | + | <StructureSection load='7t9m' size='350' frame='true' side='right' caption='Thyrotropin Receptor 7T9M' scene='95/954073/The_residues_of_p10_peptide/4'> |
This is a default text for your page '''Jackson Langford/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page '''Jackson Langford/Sandbox 1'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
[[Image:All_TSHR.png|400 px|right|thumb|Figure 1. The coolest image of this protein EVAH!]] | [[Image:All_TSHR.png|400 px|right|thumb|Figure 1. The coolest image of this protein EVAH!]] | ||
== Function == | == Function == | ||
| + | These are the residues of the P10 peptide that are largely responsible for the transduction of signal from the extra cellular domain through the transmembrane domain of the thyrotropin receptor. The thyrotropin receptor can be activated by various autoantibodies. | ||
[http://https://pubmed.ncbi.nlm.nih.gov/29771756/Thyrotropin Receptor Antibodies] | [http://https://pubmed.ncbi.nlm.nih.gov/29771756/Thyrotropin Receptor Antibodies] | ||
| - | Dr. Macbeth studied an enzyme called debranching enzyme and determined its structure through X-ray crystallography.<ref name="Ransey">PMID:28504306</ref> | ||
| - | |||
== Disease == | == Disease == | ||
| - | < | + | Dr. Macbeth studied an enzyme called debranching enzyme and determined its structure through X-ray crystallography.<ref name="Ransey">PMID:28504306</ref>. There is no real relevance to the thyrotropin receptor with this research though. |
== Relevance == | == Relevance == | ||
== Structural highlights == | == Structural highlights == | ||
| + | The residues of the p10 peptide are shown in white spheres. | ||
| + | <scene name='95/954073/The_residues_of_p10_peptide/5'>Here we can take a view from further away from the thyrotropin receptor.</scene><ref name = "Faust">PMID:35940205</ref> | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| Line 18: | Line 19: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | <references/> | ||
==Student Contributors== | ==Student Contributors== | ||
| - | <references/> | ||
Current revision
HUMAN THYROTROPIN RECEPTOR
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Faust B, Billesbolle CB, Suomivuori CM, Singh I, Zhang K, Hoppe N, Pinto AFM, Diedrich JK, Muftuoglu Y, Szkudlinski MW, Saghatelian A, Dror RO, Cheng Y, Manglik A. Autoantibody mimicry of hormone action at the thyrotropin receptor. Nature. 2022 Aug 8. pii: 10.1038/s41586-022-05159-1. doi:, 10.1038/s41586-022-05159-1. PMID:35940205 doi:http://dx.doi.org/10.1038/s41586-022-05159-1