Sandbox Reserved 1777
From Proteopedia
(Difference between revisions)
| (188 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
{{Template:CH462_Biochemistry_II_2023}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Template:CH462_Biochemistry_II_2023}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | <StructureSection load='7pui' size='350' side='right' caption='SHOC2-PP1C-MRAS (PDB entry [[ | + | <StructureSection load='7pui' size='350' side='right' caption='SHOC2-PP1C-MRAS. SHOC2(blue) PP1C(orange) MRAS (green) and metal ions (green and gray) (PDB entry [[7upi]]).' scene='95/952704/Smpcolored/1'> |
=Introduction= | =Introduction= | ||
| - | SHOC2-PP1C-MRAS is a | + | SHOC2-PP1C-MRAS is a regulatory protein and enzyme complex that is involved in regulating cell proliferation and division <Ref name='Astrain'>Bernal Astrain G, Nikolova M, Smith MJ. Functional diversity in the RAS subfamily of small GTPases. Biochem Soc Trans. 2022 Apr 29;50(2):921-933. doi: 10.1042/BST20211166. [https://doi.org/10.1042/BST20211166. DOI:10.1042/BST20211166] </Ref>. This complex regulates the vast RAS-MAPK signaling pathway through control of the initial RAF MAPKKK. This pathway is initially activated by the binding of a growth factor to a [https://www.mechanobio.info/what-is-mechanosignaling/what-are-small-gtpases/what-are-ras-gtpases/. GTPase] which initiates intracellular RAS activation, such as |
| + | [https://www.frontiersin.org/articles/10.3389/fonc.2019.01088/full#:~:text=In%20human%20cells%2C%20three%20closely,proliferation%20and%20survival%20among%20others HRAS, NRAS, or KRAS]. RAS-GTPases are a family of monomeric G-proteins that function as molecular switches and are key in regulating parts of signaling cascades. The RAS molecular switch alternates between binding GTP for the active state and GDP to become inactive <ref name="Astrain" />. | ||
| + | [[Image:Screen Shot 2023-04-16 at 7.30.09 PM.jpeg|650px|thumb|<font size="2"><div style="text-align: center;">'''Figure 1'''. Schematic representation of RAS/RAF/MEK/ERK pathway after assembly of SHOC2-PP1C-MRAS complex. </div></font>]] | ||
| + | |||
| + | After activation via an extracellular growth factor, the RAS-GTPase enzyme binds GTP, which recruits RAF to the cell membrane. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6311149/#:~:text=Full%20activation%20of%20Raf%20requires,plasma%20membrane%20(Roy%20et%20al RAF ]is a kinase that stimulates a signaling cascade by phosphorylation of MAPK (also known as MEK in mammals), which activates downstream proteins such as ERK1 and ERK2<Ref name='Molina'>Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol. 2006 Jan;1(1):7-9. [https://doi.org/10.1016/S1556-0864(15)31506-9. DOI:10.1016/S1556-0864(15)31506-9]. </Ref>. ERK1 and ERK2 subsequently activate nuclear transcription factors and kinases, as seen in Figure 1. The RAS/RAF/MEK/ERK pathway is a critical signaling cascade for activating transcription factors and regulating gene expression<Ref name='Li'>Li, L., Zhao, G. D., Shi, Z. et. al.The Ras/Raf/MEK/ERK signaling pathway (Figure 1) and its role in the occurrence and development of HCC. Oncology letters, 12(5), 3045–3050. [https://doi.org/10.3892/ol.2016.5110. DOI:10.3892/ol.2016.5110]. </Ref>. | ||
=Structure= | =Structure= | ||
| - | + | ==Overview== | |
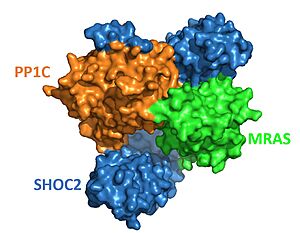
| - | == | + | [[Image:SHOC2-PP1C-MRAS Surface.JPG|300px|right|thumb|<font size="2"><div style="text-align: center;">'''Figure 2'''. Surface representation of SHOC2-PP1C-MRAS ([https://www.rcsb.org/structure/7UPI PDB 7upi]). SHOC2 (blue), PP1C (orange) and MRAS (green). </div></font>]] |
| - | [[Image:SHOC2-PP1C-MRAS Surface.JPG|300px|right|thumb|<font size=" | + | The complex combines three separate proteins, SHOC2, PP1C, and MRAS, to form the active protein (SMP complex), as seen in Figure 2<Ref name='Hauseman'>Hauseman, Z.J., Fodor, M., Dhembi, A. et al. Structure of the MRAS–SHOC2–PP1C phosphatase complex. Nature 609, 416–423 (2022). doi: 10.1038/s41586-022-05086-1. [https://doi.org/10.1038/s41586-022-05086-1. DOI:10.1038/s41586-022-05086-1]. </Ref>. In complex, each of these proteins are responsible for a different role. MRAS is the "on and off" switch; when MRAS is activated from binding GTP, it recruits SHOC2 and PP1C for complex formation. In complex, PP1C is tasked with being the catalytic domain and removing a phosphate from RAF. The last piece of the complex is SHOC2, which is responsible for holding and stabilizing MRAS and PP1C <Ref name= 'Hahn'> Kwon, J. J., & Hahn, W. C. A Leucine-Rich Repeat Protein Provides a SHOC2 the RAS Circuit: a Structure-Function Perspective. Molecular and cellular biology, 41(4), e00627-20 (2021). doi:10.1128/MCB.00627-20. [http://doi.org/10.1128/MCB.00627-20. DOI: 10.1128/MCB.00627-20]. </Ref>. The SMP complex was determined via cryo-electron microscopy as well as x-ray diffraction. The overall architecture has PP1C and MRAS bound within the concave surface of SHOC2, leaving the catalytic site of PP1C and the GTP binding cleft in MRAS exposed. |
| - | + | ||
| - | The | + | |
==SHOC2== | ==SHOC2== | ||
| + | SHOC2 is a scaffolding protein which acts as a cradle to bind PP1C and MRAS <ref name="Hauseman" />, serving as an aggregation point to enable reciprocal interactions between these three signaling proteins. <scene name='95/952705/Shoc2_structure/2'>SHOC2</scene> is a leucine rich repeat ([https://en.wikipedia.org/wiki/Leucine-rich_repeat LRR]) protein consisting of 20 consecutive <scene name='95/952706/Shoc2_structure/9'>LRR motifs</scene> containing <scene name='95/952705/Shoc2_structure/3'>leucine residues</scene>. LRR motifs form an extended β-sheet on the inner concave surface of SHOC2 with α-helices facing outward to facilitate binding of the protein complex. These LRR motifs result in a largely hydrophobic core within the concave region<ref name="Hahn" />. | ||
==PP1C== | ==PP1C== | ||
| + | <scene name='95/952705/Pp1c_structure/1'>PP1C</scene> is the catalytic domain of the phosphatase enzyme [https://www.ncbi.nlm.nih.gov/gene/5499 PP1], which removes reversible phosphorylations from signaling proteins. PP1C is a serine/threonine phosphatase involved in signaling pathways that control cell growth, division, and metabolism<Ref name= 'Aggen'> Aggen, J., Nairn, A., Chamberlin, R. Regulation of protein phosphatase-1. Chemistry & Biology 2000, 7:R13–R23. [https://www.cell.com/cell-chemical-biology/pdf/S1074-5521(00)00069-7.pdf]. </Ref>. The active site of PP1C is adjacent to a hydrophobic patch, where it binds to the N-terminal phosphoserine of RAF, its target for dephosphorylation. PP1C has phosphatase activity in the absence of the ternary complex, but it lacks intrinsic substrate selectivity<ref name="Hauseman" />. This indicates that the complex formation is necessary for PP1C's specificity for RAF. | ||
==MRAS== | ==MRAS== | ||
| + | <scene name='95/952705/Mras_structure/4'>MRAS</scene> is a monomeric GTPase and is anchored in the cell membrane by a C-terminal S-Farnesyl cysteine carboxylmethyl ester <Ref name='Zhou'> Zhou, Y., Prakash, P., Liang, H., et al. Lipid-Sorting Specificity Encoded in K-Ras Membrane Anchor Regulates Signal Output. Cell, 168(1-2), 239–251.e16 doi: 10.1016/j.cell.2016.11.059. [https://doi.org/10.1016/j.cell.2016.11.059. DOI: 10.1016/j.cell.2016.11.059]. </Ref>. When MRAS binds GTP, it becomes active, which allows MRAS to bind the rest of the complex<ref name="Hauseman" />. MRAS is a subvariant of the RAS protein and therefore shares most of its regulatory and effector interactions<Ref name= 'Young'>Young, L., Rodriguez-Viciana, P. MRAS: A Close but Understudied Member of the RAS Family. Cold Spring Harbor Perspectives in Medicine (2018). doi: 10.1101/cshperspect.a033621. [https://perspectivesinmedicine.cshlp.org/content/8/12/a033621.full.pdf+html. DOI: 0.1101/cshperspect.a033621]. </Ref>. While other RAS variants bind in complex with SHOC2 and PP1C to allow it to have phosphatase activity, MRAS binds the tightest. | ||
===Switch I and II=== | ===Switch I and II=== | ||
| + | A key feature in allowing MRAS to be in its active versus inactive state is the switches in MRAS. When MRAS is active, GTP is bound and the switches allow MRAS to be bound with PP1C. When MRAS is inactive, GDP is bound and the switches do not allow MRAS to bind to PP1C. The switches determine whether MRAS can bind to SHOC2-PP1C. The switches have to go through a conformational change to allow binding of SHOC2-PP1C to MRAS. The conformational change is needed because without it SHOC2-PP1C could bind to MRAS when MRAS is still inactive. This process would cause the SHOC2-PP1C-MRAS pathway to constantly be running. The switches and GDP/GTP help regulate this process. | ||
| - | + | This conformational change is caused by GTP replacing GDP. When GTP is bound, MRAS shifts and binds with the previously associated SHOC2-PP1C complex. When GDP is bound, <scene name='95/952705/Switch_i_and_ii_with_gdp/8'>switch II</scene> in MRAS is moved outward, which causes a <scene name='95/952705/Gdp_with_mras_and_shoc2/2'>steric clash with SHOC2</scene>. When GTP is bound, <scene name='95/952705/Switch_i_and_ii_with_gtp/5'>switch II</scene> in MRAS can form various hydrogen bonding, pi stacking and hydrophobic interactions with SHOC2<Ref name='Bonsor'>Daniel A. Bonsor, Patrick Alexander, Kelly Snead, Nicole Hartig, Matthew Drew, Simon Messing, Lorenzo I. Finci, Dwight V. Nissley, Frank McCormick, Dominic Esposito, Pablo Rodrigiguez-Viciana, Andrew G. Stephen, Dhirendra K. Simanshu. Structure of the SHOC2–MRAS–PP1C complex provides insights into RAF activation and Noonan syndrome. bioRxiv. 2022.05.10.491335. doi: 10.1101/2022.05.10.491335. [https://doi.org/10.1101/2022.05.10.491335. DOI:10.1101/2022.05.10.491335]. </Ref>. When MRAS is bound to SHOC2 and PP1C, switch I has an important role in making <scene name='95/952705/Switch_i_and_ii_with_gtp/7'>interactions with PP1C</scene>. Though it is minute, there is a change of the positioning of switch II when GDP vs GTP is bound. | |
| - | == | + | ==Stabilizing Interactions in Ternary Complex== |
| + | Once an initial signal to begin complex formation has been received, SHOC2 binds PP1C to form a binary complex. The SHOC2-PP1C complex then binds to the membrane bound MRAS. <scene name='95/952705/Shoc2_pp1c_interaction/3'>PP1C binds</scene> with the ascending loops of the SHOC2 LRR regions, and is further engaged through the N-terminal region of SHOC2 which contains the <scene name='95/952706/Shoc2_rvxf/3'>RVxF motif</scene> <Ref name= 'Jajian'>Kwon, J., Jajian, B., Bian, Y. et al. Comprehensive structure-function evaluation of the SHOC2 holophosphatase reveals disease mechanisms and therapeutic opportunities. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022. [https://aacrjournals.org/cancerres/article/82/12_Supplement/LB029/699443. DOI: 10.1158/1538-7445.AM2022-LB029]. </Ref>. The initial forming of the complex begins with SHOC2-PP1C engagement, then is completed and stabilized by the GTP-loaded MRAS binding<Ref name="Jajian" />. <scene name='95/952705/Shoc2_mras_interaction/3'>MRAS binds to SHOC2</scene> exclusively through its concave LRRs<Ref name='Kwan'>Kwon, J.J., Hajian, B., Bian, Y. et al. Structure–function analysis of the SHOC2–MRAS–PP1C holophosphatase complex. Nature 609, 408–415 (2022).doi: 10.1038/s41586-022-04928-2. [https://doi.org/10.1038/s41586-022-04928-2. DOI:10.1038/s41586-022-04928-2] </Ref>, primarily by the descending loop and strands of LRR domains 2-10. Once associated with SHOC2, <scene name='95/952705/Mras_pp1c_interaction/3'>MRAS binds to PP1C</scene>. Binding to MRAS localizes the other two proteins to the RAS signaling regions of the membrane to begin cellular signaling<ref name="Kwan" />. | ||
| + | |||
| + | ==Active Site of PP1C== | ||
| + | Once all the domains are bound, [https://en.wikipedia.org/wiki/Nucleoside_triphosphate NTpS] binds to the <scene name='95/952705/Pp1c_active_site/6'>active site of PP1C</scene>. NTpS refers to the phosphorylated serine (residue 259) located at the N-terminus of RAF. Once NTpS is bound, PP1C catalyzes the dephosphorylation of serine 259. NTpS is dephosphorylated to prevent the active dimeric RAF from inactivating and changing into its monomeric structure. | ||
| - | + | Notably, NTpS can bind to the PP1C active site without PP1C being in complex with SHOC2 and MRAS. This occurs when RAS binds to RAF, which exposes NTpS on RAF. However, for this reaction, the catalytic activity of PP1C outside of the complex is less efficient. It's hypothesized this is because hydrophobic residues directly C-terminal to the phosphorylated serine bind to the hydrophobic patch of PP1C as well as the hydrophobic SHOC2 C-terminus<ref name="Liau">PMID:35768504</ref>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | . This | + | |
| - | + | The conserved hydrophobic groove at the SHOC2 C-terminus and the hydrophobic region next to the PP1C active site neighbor each other in the structure of the complex. As a result, the hydrophobic residues downstream from NTpS bind to the hydrophobic patch of PP1C next to the active site and to the hydrophobic groove in the SHOC2 C-terminus. There is still some uncertainty as to how the substrate selectivity works, but these regions could play an essential role in the promiscuous binding of PP1C<ref name="Liau">PMID:35768504</ref>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | =Activation of RAF= | ||
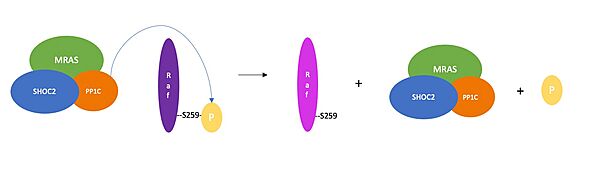
| + | [[Image:Screen Shot 2023-04-10 at 4.34.56 PM.jpeg|600px|thumb|<font size="2"><div style="text-align: center;">'''Figure 3'''. Schematic representation of RAF activation. PP1C removes an inhibitory phosphate group from RAF, which results in RAF activation and activation of the RAF/MAPK signaling cascade. </div></font>]] | ||
| + | Once the SMP complex comes together, it plays a key role in regulating the activation of the RAF/MAPK pathway. PP1C, enhanced through interactions with SHOC2 and MRAS, removes a phosphate group from serine residue 259 (also known as NTpS) of RAF (Figure 3). The removal of the phosphate group triggers activation of RAF, a serine/threonine kinase, which leads to the phosphorylation and activation of MAPK <ref name="Molina" />. This causes the activation of downstream proteins ERK1 and ERK2, which are responsible for activating nuclear transcription factors such as Elk-1, c-Ets1, c-Ets2, and MNK1, which result in the transcription of genes that code for proteins which promote cell proliferation and differentiation. <Ref name='Lavoie'>Lavoie, H., Therrien, M. Structural keys unlock RAS–MAPK cellular signaling pathway. Nature 609, 248-249 (2022). doi: 10.1038/d41586-022-02189-7. [https://doi.org/10.1038/d41586-022-02189-7. DOI:10.1038/d41586-022-02189-7]. </Ref>. | ||
=Implications= | =Implications= | ||
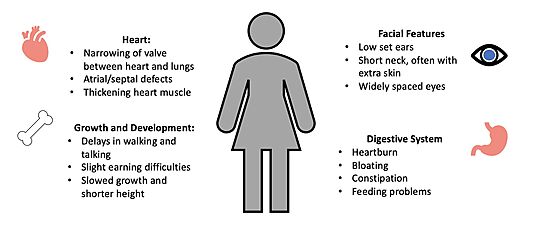
| - | + | [[Image:Noonan syndrome figure.jpg|550px|right|thumb|<font size="2"><div style="text-align: center;">'''Figure 4'''. Common Symptoms of Noonan Syndrome. </div></font>]] | |
| - | The | + | The SHOC2-PP1C-MRAS complex's key role in the regulation of the MAPK-RAF pathway means that minor changes in its structure or function can have drastic biological consequences. Unregulated activation of the MAPK pathway is the cause of several human cancers due to unchecked cell division and proliferation<ref name="Kwan" />. Mutations that stabilize the interactions of the SMP complex enhance PP1C phosphatase activity <Ref name="Jajian" />, leading to increased RAF signaling and accelerated cell division. Most SHOC2 or PP1C mutations alter residues that are the direct contact points, stabilizing the interaction between the two proteins. Mutations to MRAS result in persistent binding of GTP, leading to consistent activation of the cell signaling pathway<ref name="Kwan" />. |
| + | The unregulated MAPK pathway is also responsible for a multitude of developmental disorders commonly known as [https://dceg.cancer.gov/research/what-we-study/rasopathies#:~:text=RASopathies%20are%20a%20group%20of,to%20grow%20and%20work%20properly. RASopathies] <Ref name="Lavoie" />. One RASopathy caused by the mutation of a RAF kinase is known as [https://www.mayoclinic.org/diseases-conditions/noonan-syndrome/symptoms-causes/syc-20354422 Noonan Syndrome] (NS), which enhances complex formation by stabilizing the interactions of each member <ref name="Kwan" />. NS is a genetic disorder that can prevent normal development during the neonatal period, leading to difficulties with feeding and a failure to thrive<Ref name= 'van der Burgt'> van der Burgt, I. Noonan syndrome. Orphanet J Rare Dis 2, 4 (2007). doi: 10.1186/1750-1172-2-4 [https://doi.org/10.1186/1750-1172-2-4. DOI: 10.1186/1750-1172-2-4]. </Ref>. The characteristic features of NS become more evident during infancy and childhood. NS patients often have atypical facial appearance, short stature, heart defects, and other physical problems (Figure 4)<ref name="van der Burgt" />. | ||
| + | </StructureSection> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ 1.0 1.1 Bernal Astrain G, Nikolova M, Smith MJ. Functional diversity in the RAS subfamily of small GTPases. Biochem Soc Trans. 2022 Apr 29;50(2):921-933. doi: 10.1042/BST20211166. DOI:10.1042/BST20211166
- ↑ 2.0 2.1 Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol. 2006 Jan;1(1):7-9. DOI:10.1016/S1556-0864(15)31506-9.
- ↑ Li, L., Zhao, G. D., Shi, Z. et. al.The Ras/Raf/MEK/ERK signaling pathway (Figure 1) and its role in the occurrence and development of HCC. Oncology letters, 12(5), 3045–3050. DOI:10.3892/ol.2016.5110.
- ↑ 4.0 4.1 4.2 4.3 Hauseman, Z.J., Fodor, M., Dhembi, A. et al. Structure of the MRAS–SHOC2–PP1C phosphatase complex. Nature 609, 416–423 (2022). doi: 10.1038/s41586-022-05086-1. DOI:10.1038/s41586-022-05086-1.
- ↑ 5.0 5.1 Kwon, J. J., & Hahn, W. C. A Leucine-Rich Repeat Protein Provides a SHOC2 the RAS Circuit: a Structure-Function Perspective. Molecular and cellular biology, 41(4), e00627-20 (2021). doi:10.1128/MCB.00627-20. DOI: 10.1128/MCB.00627-20.
- ↑ Aggen, J., Nairn, A., Chamberlin, R. Regulation of protein phosphatase-1. Chemistry & Biology 2000, 7:R13–R23. [1].
- ↑ Zhou, Y., Prakash, P., Liang, H., et al. Lipid-Sorting Specificity Encoded in K-Ras Membrane Anchor Regulates Signal Output. Cell, 168(1-2), 239–251.e16 doi: 10.1016/j.cell.2016.11.059. DOI: 10.1016/j.cell.2016.11.059.
- ↑ Young, L., Rodriguez-Viciana, P. MRAS: A Close but Understudied Member of the RAS Family. Cold Spring Harbor Perspectives in Medicine (2018). doi: 10.1101/cshperspect.a033621. DOI: 0.1101/cshperspect.a033621.
- ↑ Daniel A. Bonsor, Patrick Alexander, Kelly Snead, Nicole Hartig, Matthew Drew, Simon Messing, Lorenzo I. Finci, Dwight V. Nissley, Frank McCormick, Dominic Esposito, Pablo Rodrigiguez-Viciana, Andrew G. Stephen, Dhirendra K. Simanshu. Structure of the SHOC2–MRAS–PP1C complex provides insights into RAF activation and Noonan syndrome. bioRxiv. 2022.05.10.491335. doi: 10.1101/2022.05.10.491335. DOI:10.1101/2022.05.10.491335.
- ↑ 10.0 10.1 10.2 Kwon, J., Jajian, B., Bian, Y. et al. Comprehensive structure-function evaluation of the SHOC2 holophosphatase reveals disease mechanisms and therapeutic opportunities. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022. DOI: 10.1158/1538-7445.AM2022-LB029.
- ↑ 11.0 11.1 11.2 11.3 11.4 Kwon, J.J., Hajian, B., Bian, Y. et al. Structure–function analysis of the SHOC2–MRAS–PP1C holophosphatase complex. Nature 609, 408–415 (2022).doi: 10.1038/s41586-022-04928-2. DOI:10.1038/s41586-022-04928-2
- ↑ 12.0 12.1 Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J. Structural basis for SHOC2 modulation of RAS signalling. Nature. 2022 Jun 29. pii: 10.1038/s41586-022-04838-3. doi:, 10.1038/s41586-022-04838-3. PMID:35768504 doi:http://dx.doi.org/10.1038/s41586-022-04838-3
- ↑ 13.0 13.1 Lavoie, H., Therrien, M. Structural keys unlock RAS–MAPK cellular signaling pathway. Nature 609, 248-249 (2022). doi: 10.1038/d41586-022-02189-7. DOI:10.1038/d41586-022-02189-7.
- ↑ 14.0 14.1 van der Burgt, I. Noonan syndrome. Orphanet J Rare Dis 2, 4 (2007). doi: 10.1186/1750-1172-2-4 DOI: 10.1186/1750-1172-2-4.