Sandbox Reserved 1768
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 3: | Line 3: | ||
== Introduction == | == Introduction == | ||
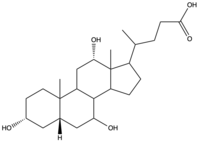
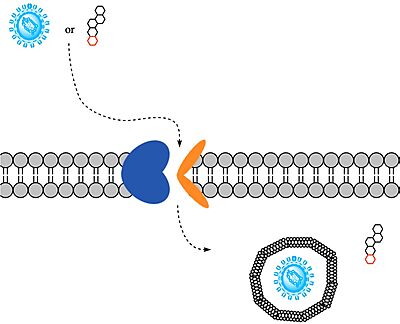
| - | Sodium-taurocholate Co-transporting Polypeptide (NTCP) is a member of the solute carrier membrane transport protein family. It is found within the membrane of [[Image:Bile Salt.png|200 px|right|thumb|Figure 1. Structure of [https://en.wikipedia.org/wiki/Cholic_acid Cholic Acid], an example of a bile acid.]][https://en.wikipedia.org/wiki/Hepatocyte hepatocytes], and its primary role is to facilitate the transport of [https://en.wikipedia.org/wiki/Bile_acid bile salts] into hepatocytes from the bloodstream (Figure 1).<Ref name="Goutam"> Goutam K, Ielasi FS, Pardon E, Steyaert J, Reyes N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022 Jun;606(7916):1015-1020. [https://dx.doi.org/10.1038/s41586-022-04723-z DOI: 10.1038/s41586-022-04723-z]. </Ref> This is important because 90% of human bile salts are recycled daily, so the function of NTCP is critical in providing bile acids to solubilize fats for digestion. In addition to transporting bile acids into the cytoplasm of hepatocytes, NTCP also serves as an entry point receptor for [https://en.wikipedia.org/wiki/Hepatitis_B Hepatitis B (HBV)] and [https://en.wikipedia.org/wiki/Hepatitis_D Hepatitis D (HDV)] viruses (Figure 2).<Ref name="Asami"> Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. [https://dx.doi.org/10.1038/s41586-022-04845-4 DOI: 10.1038/s41586-022-04845-4]. </Ref> [[Image:Ntcpmech.jpg|400 px|thumb|Figure 2. Overall NTCP mechanism. Bile salts and HBV/HDV viruses bind to NTCP. NTCP transports bile salts through an inner pore, releasing the bile salt into the cytoplasm. Transport of viral particles occurs via endocytosis of NTCP.]] | + | Sodium-taurocholate Co-transporting Polypeptide (NTCP) is a member of the solute carrier membrane transport protein family. It is found within the membrane of [[Image:Bile Salt.png|200 px|right|thumb|Figure 1. Structure of [https://en.wikipedia.org/wiki/Cholic_acid Cholic Acid], an example of a bile acid.]][https://en.wikipedia.org/wiki/Hepatocyte hepatocytes], and its primary role is to facilitate the transport of [https://en.wikipedia.org/wiki/Bile_acid bile salts] into hepatocytes from the bloodstream (Figure 1).<Ref name="Goutam"> Goutam K, Ielasi FS, Pardon E, Steyaert J, Reyes N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022 Jun;606(7916):1015-1020. [https://dx.doi.org/10.1038/s41586-022-04723-z DOI: 10.1038/s41586-022-04723-z]. </Ref> This is important because 90% of human bile salts are recycled daily, so the function of NTCP is critical in providing bile acids to solubilize fats for digestion. In addition to transporting bile acids into the cytoplasm of hepatocytes, NTCP also serves as an entry point receptor for [https://en.wikipedia.org/wiki/Hepatitis_B Hepatitis B (HBV)] and [https://en.wikipedia.org/wiki/Hepatitis_D Hepatitis D (HDV)] viruses (Figure 2).<Ref name="Asami"> Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. [https://dx.doi.org/10.1038/s41586-022-04845-4 DOI: 10.1038/s41586-022-04845-4]. </Ref> [[Image:Ntcpmech.jpg|400 px|thumb|Figure 2. Overall NTCP mechanism. Bile salts and HBV/HDV [https://en.wikipedia.org/wiki/Virus viruses] bind to NTCP. NTCP transports bile salts through an inner pore, releasing the bile salt into the cytoplasm. Transport of viral particles occurs via endocytosis of NTCP.]] |
== Structure == | == Structure == | ||
| - | NTCP is a transmembrane protein found in hepatocyte cells. It consists of nine <scene name='95/952697/Ntcp_open-pore_state/22'>transmembrane alpha helices</scene>, with the N-terminus located on the extracellular side of the plasma membrane and the C-terminus located on the intracellular side (Figure 3). The transmembrane helices are connected by short loops as well as extracellular and intracellular alpha helices that lie nearly parallel to the membrane.<ref name="Asami" /> Structures of NTCP were determined by [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryogenic electron microscopy (Cryo-EM)] of NTCP in complex with antibodies or nanobodies, which served to stabilize two known conformational states of NTCP. <ref name="Goutam" /> <ref name="Asami" /> <Ref name="Park"> Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022 Jun;606(7916):1027-1031. [https://dx.doi.org/10.1038/s41586-022-04857-0 DOI: 10.1038/s41586-022-04857-0]. </Ref> <ref name="Liu" /> These complexed antibodies/nanobodies have been removed from all structures shown on this page. | + | NTCP is a transmembrane protein found in hepatocyte cells. It consists of nine <scene name='95/952697/Ntcp_open-pore_state/22'>transmembrane alpha helices</scene>, with the N-terminus located on the extracellular side of the plasma membrane and the C-terminus located on the intracellular side (Figure 3). The transmembrane helices are connected by short loops as well as extracellular and intracellular alpha helices that lie nearly parallel to the membrane.<ref name="Asami" /> Structures of NTCP were determined by [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryogenic electron microscopy (Cryo-EM)] of NTCP in complex with [https://en.wikipedia.org/wiki/Antibody antibodies] or [https://en.wikipedia.org/wiki/Single-domain_antibody nanobodies], which served to stabilize two known conformational states of NTCP. <ref name="Goutam" /> <ref name="Asami" /> <Ref name="Park"> Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022 Jun;606(7916):1027-1031. [https://dx.doi.org/10.1038/s41586-022-04857-0 DOI: 10.1038/s41586-022-04857-0]. </Ref> <ref name="Liu" /> These complexed antibodies/nanobodies have been removed from all structures shown on this page. |
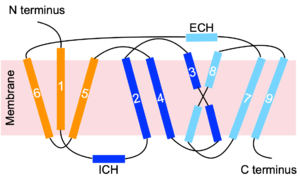
[[Image:Topology_picture.png|300 px|thumb|Figure 3. NTCP topology. Transmembrane helices are numbered. The panel domain is shown in orange. The core domain is shown in two shades of blue. Light and dark blue shades represent different helix bundles of the core domain, which together demonstrate pseudo two-fold symmetry.]] | [[Image:Topology_picture.png|300 px|thumb|Figure 3. NTCP topology. Transmembrane helices are numbered. The panel domain is shown in orange. The core domain is shown in two shades of blue. Light and dark blue shades represent different helix bundles of the core domain, which together demonstrate pseudo two-fold symmetry.]] | ||
| Line 15: | Line 15: | ||
** Formed by transmembrane helices TM1, TM5, and TM6. | ** Formed by transmembrane helices TM1, TM5, and TM6. | ||
*<b><font color="#0040e0">Core domain</font></b>: <scene name='95/952697/Ntcp_open-pore_state/24'>TM helices 2-4, 7-9</scene> | *<b><font color="#0040e0">Core domain</font></b>: <scene name='95/952697/Ntcp_open-pore_state/24'>TM helices 2-4, 7-9</scene> | ||
| - | **Formed by the packing of a helix bundle of <b><font color="blue">TM2, TM3, and TM4</font></b> with another helix bundle of <b><font color="skyblue">TM7, TM8, and TM9</font></b> (Figure 3). These <scene name='95/952697/Ntcp_open-pore_state/ | + | **Formed by the packing of a helix bundle of <b><font color="blue">TM2, TM3, and TM4</font></b> with another helix bundle of <b><font color="skyblue">TM7, TM8, and TM9</font></b> (Figure 3). These <scene name='95/952697/Ntcp_open-pore_state/41'>two helix bundles</scene> are related by pseudo two-fold symmetry.<Ref name="Qi"> Qi X, Li W. Unlocking the secrets to human NTCP structure. Innovation (Camb). 2022 Aug 1;3(5):100294. [https://dx.doi.org/10.1016/j.xinn.2022.100294 DOI: 10.1016/j.xinn.2022.100294]. </Ref> |
=== Proline/Glycine Hinge === | === Proline/Glycine Hinge === | ||
| - | <scene name='95/952697/Ntcp_open-pore_state/ | + | <scene name='95/952697/Ntcp_open-pore_state/45'>Glycine and proline residues</scene> in the <scene name='95/952697/Ntcp_open-pore_state/43'>connecting loops</scene> and <scene name='95/952697/Ntcp_open-pore_state/42'>extra- (ECH) and intracellular helices (ICH)</scene> (Figure 3) act as hinges between the <scene name='95/952697/Ntcp_open-pore_state/44'>open pore state</scene> and <scene name='95/952697/Ntcp_inward_facing_state/5'>inward-facing states</scene> in the mechanism of bile salt uptake. This flexibility allows separation of the core and panel domains, creating a pore open to the extracellular space and exposing critical Na+ binding sites. Once substrate binds the open-pore state, this hinge allows the transition to close this pore relative to the extracellular side and open to the cytoplasmic side, thus allowing release of substrate into the cell.<ref name = "Goutam" /> |
=== Sodium Binding Sites === | === Sodium Binding Sites === | ||
| - | To transport a single bile salt from the blood to the cytoplasm of the hepatocyte, two sodium ions must bind to NTCP in the open-pore state, using two <scene name='95/952697/Bothnabindingsites/ | + | To transport a single bile salt from the blood to the cytoplasm of the hepatocyte, <scene name='95/952697/Bothnabindingsites/4'>two sodium ions</scene> must bind to NTCP in the open-pore state, using two <scene name='95/952697/Bothnabindingsites/7'>sodium binding sites</scene>.<Ref name="Liu"> Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. [https://dx.doi.org/10.1038/s41422-022-00680-4 DOI: 10.1038/s41422-022-00680-4]. </Ref> The residues in the <scene name='95/952697/Ntcp_complex_sodiumsites/14'>first sodium binding site</scene> include S105, N106, T123, and E257. The residues in the <scene name='95/952697/Ntcp_complex_sodiumsites/13'>second sodium binding site</scene> include Q68 and Q261. Mutations to these significant residues inhibit the binding of sodium ions, and consequently, inhibit the transport of bile salts by NTCP.<ref name = "Liu" /> [https://en.wikipedia.org/wiki/Active_transport#Secondary_active_transport Secondary active transport] is used, as the transport of bile acids into the cell is thermodynamically unfavorable, but is coupled with the favorable transport of two sodium into into the cell to make the overall process thermodynamically favorable.<ref name = "Goutam" /> When the bile salts are released into the cell, NTCP transitions to the <scene name='95/952697/Ntcp_inward_facing_state/3'>inward facing state</scene>, where the pore is now closed to the extracellular side. |
== Function == | == Function == | ||
| Line 27: | Line 27: | ||
=== Mechanism of Bile Salt Uptake === | === Mechanism of Bile Salt Uptake === | ||
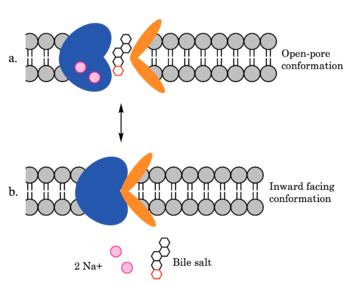
[[Image:ntcpmechanismoverall.png|350 px|right|thumb| Figure 4. Mechanism of bile salt uptake by NTCP. A bile salt and two Na+ ions bind to NTCP in the open-pore state. Transition to the inward-facing state allows release of the bile salt and ions into the cytoplasm.]] | [[Image:ntcpmechanismoverall.png|350 px|right|thumb| Figure 4. Mechanism of bile salt uptake by NTCP. A bile salt and two Na+ ions bind to NTCP in the open-pore state. Transition to the inward-facing state allows release of the bile salt and ions into the cytoplasm.]] | ||
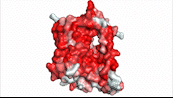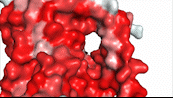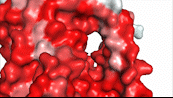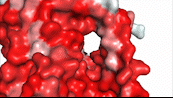
| - | NTCP utilizes secondary active transport to uptake bile salts from blood plasma into the cytoplasm of liver cells (Figure 4). The transport of one bile salt is driven by the downhill transport of 2 Na+ ions along sodium’s electrochemical gradient. | + | NTCP utilizes secondary active transport to uptake bile salts from blood plasma into the cytoplasm of liver cells (Figure 4). The transport of one bile salt is driven by the downhill transport of 2 Na+ ions along sodium’s electrochemical gradient. Two bile salts and two Na+ ions bind to the <scene name='95/952697/Ntcp_open-pore_state_surface/4'>open-pore state</scene> (Figure 4a), but only one bile salt (along with two Na+ ions) is released into the cytoplasm.<ref name="Liu" /> After all substrates are bound, the conformational change from the open-pore state to the inward facing state is driven by the energetics of the favorable transport of Na+ ions.<ref name="Liu" /> The <scene name='95/952697/Ntcp_inward_facing_state/3'>inward facing state</scene> (Figure 4b) allows release of one bile salt and two Na+ ions into the cytoplasm. In this conformation, the pore closes off relative to the extracellular side and opens to the cytoplasmic side.<ref name="Asami" /> Afterwards, the remaining bile salt bound to NTCP shifts to the position the previous bile salt occupied, and the process repeats itself.<ref name = "Liu" /> The inherent amiphipathicity of bile acids allows passage through NTCP’s amphipathic pore (Figure 5)[[Image:Hydro_NEWEST_AdobeExpress_(1).gif|400 px|right|thumb|Figure 5. Amphipathic pore of NTCP highlighting hydrophobic residues (red) and hydrophilic residues (white). (PDB: [https://www.rcsb.org/structure/7PQQ 7PQQ])]] |
=== Mechanism of HBV/HDV Infection === | === Mechanism of HBV/HDV Infection === | ||
After binding to NTCP in the <scene name='95/952697/Ntcp_open-pore_state/26'>open-pore state</scene>, the viruses remain bound until low bile salt levels in the blood shift equilibria enough that [https://en.wikipedia.org/wiki/Endocytosis endocytosis] of the virus occurs. Once inside the cell, the viral genetic information is released. | After binding to NTCP in the <scene name='95/952697/Ntcp_open-pore_state/26'>open-pore state</scene>, the viruses remain bound until low bile salt levels in the blood shift equilibria enough that [https://en.wikipedia.org/wiki/Endocytosis endocytosis] of the virus occurs. Once inside the cell, the viral genetic information is released. | ||
| - | The exact mechanism of how HBV and HDV bind to NTCP is not certain, <scene name='95/952696/Residues_84-87_and_157-165_new/1'>two critical sites</scene> on NTCP for HBV/HDV binding have been identified: residues <scene name='95/952696/Residues_84-87_new/1'>84-87</scene> and <scene name='95/952696/Residues_157-165_new/1'>157-165</scene>. An additional [https://en.wikipedia.org/wiki/Single-nucleotide_polymorphism single-nucleotide polymorphism] was discovered in East Asia involving <scene name='95/952696/Residue_267_new/15'>residue 267</scene> being mutated from serine to phenylalanine. This mutation prevented HBV/HDV infection presumably by blocking the binding site in the pore of NTCP. Another mutation, replacing <scene name='95/952696/Leucine_residues/2'>L27, L31, and L35</scene> with tryptophan residues prevents HBV/HDV infection. Further zooming in on <scene name='95/952696/Leucine_residues_zoomed/ | + | The exact mechanism of how HBV and HDV bind to NTCP is not certain, <scene name='95/952696/Residues_84-87_and_157-165_new/1'>two critical sites</scene> on NTCP for HBV/HDV binding have been identified: residues <scene name='95/952696/Residues_84-87_new/1'>84-87</scene> and <scene name='95/952696/Residues_157-165_new/1'>157-165</scene>. An additional [https://en.wikipedia.org/wiki/Single-nucleotide_polymorphism single-nucleotide polymorphism] was discovered in East Asia involving <scene name='95/952696/Residue_267_new/15'>residue 267</scene> being mutated from serine to phenylalanine. This mutation prevented HBV/HDV infection presumably by blocking the binding site in the pore of NTCP. Another mutation, replacing <scene name='95/952696/Leucine_residues/2'>L27, L31, and L35</scene> with tryptophan residues prevents HBV/HDV infection. Further zooming in on <scene name='95/952696/Leucine_residues_zoomed/2'>these residues</scene>, it is probable that this mutation blocks the preS1 binding site of HBV/HDV. [https://en.wikipedia.org/wiki/Myristoylation Myristoylation] of the HBV/HDV capsid is also vital for recognition by NTCP, as well as residues 8-17 on HBV/HDV (sequence: NPLGFFPDHQ)<ref name="Park"/>. Two mechanisms have been proposed for how HBV/HDV binds to NTCP. The first mechanism involves binding of the myristoyl group to the host cell membrane, while residues 8-17 interact with NTCP residues <scene name='95/952696/Residues_157-165_new/1'>157-165</scene>. The second mechanism involves binding of the myristoyl group with residues 157-165 in the pore.<Ref name="Zhang"> Zhang X, Zhang Q, Peng Q, Zhou J, Liao L, Sun X, Zhang L, Gong T. Hepatitis B virus preS1-derived lipopeptide functionalized liposomes for targeting of hepatic cells. Biomaterials. 2014 Jul;35(23):6130-41. [https://dx.doi.org/10.1016/j.biomaterials.2014.04.037 DOI: 10.1016/j.biomaterials.2014.04.037]. </Ref> After successful binding via one of these two proposed mechanisms, the virus is eventually endocytosed as described above. |
== Medical Relevance == | == Medical Relevance == | ||
Bile salts are derived from [https://en.wikipedia.org/wiki/Cholesterol cholesterol], and they serve an important role in the mechanical digestion of fats and ultimately facilitate the chemical digestion of lipids.<Ref name="Patton"> Patton JS, Carey MC. Watching fat digestion. Science. 1979 Apr 13;204(4389):145-8. [https://dx.doi.org/10.1126/science.432636 DOI: 10.1126/science.432636]. </Ref> Their [https://en.wikipedia.org/wiki/Amphiphile amphipathicity] allows them to solubilize hydrophobic fats for transport in aqueous bodily fluids.<ref name = "Patton" /> Without bile salts, fats would spontaneously separate out of the aqueous solution in the duodenum and would not be accessible to [https://en.wikipedia.org/wiki/Pancreatic_lipase_family#Human_pancreatic_lipase pancreatic lipase] for breakdown. Proper fat digestion requires both pancreatic lipase and bile; thus, NTCP's function in recycling bile salts is critical.<ref name = "Patton" /> Additionally, bile acids play a major role in the regulation of lipid and energy metabolism. In mice, targeting NTCP-mediated bile acid was shown to treat obesity and obesity-related [http://www.example.com hepatosteatosis] through the simultaneous dampening of intestinal fat absorption and increasing energy expenditure.<Ref name="Donkers"> Donkers JM, Kooijman S, Slijepcevic D, Kunst RF, Roscam Abbing RL, Haazen L, de Waart DR, Levels JH, Schoonjans K, Rensen PC, Oude Elferink RP, van de Graaf SF. NTCP deficiency in mice protects against obesity and hepatosteatosis. JCI Insight. 2019 Jun 25;5(14):e127197. [https://dx.doi.org/10.1172/jci.insight.127197 DOI: 10.1172/jci.insight.127197]. </Ref> | Bile salts are derived from [https://en.wikipedia.org/wiki/Cholesterol cholesterol], and they serve an important role in the mechanical digestion of fats and ultimately facilitate the chemical digestion of lipids.<Ref name="Patton"> Patton JS, Carey MC. Watching fat digestion. Science. 1979 Apr 13;204(4389):145-8. [https://dx.doi.org/10.1126/science.432636 DOI: 10.1126/science.432636]. </Ref> Their [https://en.wikipedia.org/wiki/Amphiphile amphipathicity] allows them to solubilize hydrophobic fats for transport in aqueous bodily fluids.<ref name = "Patton" /> Without bile salts, fats would spontaneously separate out of the aqueous solution in the duodenum and would not be accessible to [https://en.wikipedia.org/wiki/Pancreatic_lipase_family#Human_pancreatic_lipase pancreatic lipase] for breakdown. Proper fat digestion requires both pancreatic lipase and bile; thus, NTCP's function in recycling bile salts is critical.<ref name = "Patton" /> Additionally, bile acids play a major role in the regulation of lipid and energy metabolism. In mice, targeting NTCP-mediated bile acid was shown to treat obesity and obesity-related [http://www.example.com hepatosteatosis] through the simultaneous dampening of intestinal fat absorption and increasing energy expenditure.<Ref name="Donkers"> Donkers JM, Kooijman S, Slijepcevic D, Kunst RF, Roscam Abbing RL, Haazen L, de Waart DR, Levels JH, Schoonjans K, Rensen PC, Oude Elferink RP, van de Graaf SF. NTCP deficiency in mice protects against obesity and hepatosteatosis. JCI Insight. 2019 Jun 25;5(14):e127197. [https://dx.doi.org/10.1172/jci.insight.127197 DOI: 10.1172/jci.insight.127197]. </Ref> | ||
| - | Insight into NTCP's structure and function has implications for therapeutic treatment of HBV/HDV infection. The overlap in NTCP binding sites for bile salts and HBV/HDV viruses presents therapeutic issues in blocking HBV/HDV infection without inhibiting normal bile acid uptake. For example, mycludex B is a current first-class inhibitor of HDV infection, but suffers the drawback of hindering bile acid uptake. Potentially overcoming this, the inhibitory effect of antibody Nb87 on myr-preS1 binding shows potential for therapeutics that stabilize NTCP inward facing state as an [https://en.wikipedia.org/wiki/Allosteric_regulation allosteric inhibitor] of viral cell entry. <Ref name="Zhang" /> With Nb87 bound, HBV/HDV infection was reduced, although transport of bile salts persisted. Another study has identified a [https://en.wikipedia.org/wiki/Ciclosporin ciclosporin] derivative that has the capability to prevent HBV binding while still allowing NTCP to transport bile salts. <Ref name="Shimura"> Shimura S, Watashi K, Fukano K, Peel M, Sluder A, Kawai F, Iwamoto M, Tsukuda S, Takeuchi JS, Miyake T, Sugiyama M, Ogasawara Y, Park SY, Tanaka Y, Kusuhara H, Mizokami M, Sureau C, Wakita T. Cyclosporin derivatives inhibit hepatitis B virus entry without interfering with NTCP transporter activity. J Hepatol. 2017 Apr;66(4):685-692. doi: 10.1016/j.jhep.2016.11.009. [https://dx.doi.org/10.1016/j.jhep.2016.11.009 DOI: 10.1016/j.jhep.2016.11.009]. </Ref> This small molecule is the first to successfully prevent infection against multiple HBV genotypes without hindering normal NTCP activity, highlighting the fact that anti-HBV activity can remain separate from NTCP function.<ref name="Shimura"/> Future studies will work to identify more effective and safe ways to prevent infection without hindering normal bile acid uptake. | + | Insight into NTCP's structure and function has implications for therapeutic treatment of HBV/HDV infection. The overlap in NTCP binding sites for bile salts and HBV/HDV viruses presents therapeutic issues in blocking HBV/HDV infection without inhibiting normal bile acid uptake. For example, [https://en.wikipedia.org/wiki/Bulevirtide mycludex B] is a current first-class inhibitor of HDV infection, but suffers the drawback of hindering bile acid uptake. Potentially overcoming this, the inhibitory effect of antibody Nb87 on myr-preS1 binding shows potential for therapeutics that stabilize NTCP inward facing state as an [https://en.wikipedia.org/wiki/Allosteric_regulation allosteric inhibitor] of viral cell entry. <Ref name="Zhang" /> With Nb87 bound, HBV/HDV infection was reduced, although transport of bile salts persisted. Another study has identified a [https://en.wikipedia.org/wiki/Ciclosporin ciclosporin] derivative that has the capability to prevent HBV binding while still allowing NTCP to transport bile salts. <Ref name="Shimura"> Shimura S, Watashi K, Fukano K, Peel M, Sluder A, Kawai F, Iwamoto M, Tsukuda S, Takeuchi JS, Miyake T, Sugiyama M, Ogasawara Y, Park SY, Tanaka Y, Kusuhara H, Mizokami M, Sureau C, Wakita T. Cyclosporin derivatives inhibit hepatitis B virus entry without interfering with NTCP transporter activity. J Hepatol. 2017 Apr;66(4):685-692. doi: 10.1016/j.jhep.2016.11.009. [https://dx.doi.org/10.1016/j.jhep.2016.11.009 DOI: 10.1016/j.jhep.2016.11.009]. </Ref> This small molecule is the first to successfully prevent infection against multiple HBV genotypes without hindering normal NTCP activity, highlighting the fact that anti-HBV activity can remain separate from NTCP function.<ref name="Shimura"/> Future studies will work to identify more effective and safe ways to prevent infection without hindering normal bile acid uptake. |
Current revision
Sodium-taurocholate Co-transporting Polypeptide
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Goutam K, Ielasi FS, Pardon E, Steyaert J, Reyes N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022 Jun;606(7916):1015-1020. DOI: 10.1038/s41586-022-04723-z.
- ↑ 2.0 2.1 2.2 2.3 Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. DOI: 10.1038/s41586-022-04845-4.
- ↑ 3.0 3.1 Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022 Jun;606(7916):1027-1031. DOI: 10.1038/s41586-022-04857-0.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. DOI: 10.1038/s41422-022-00680-4.
- ↑ Qi X, Li W. Unlocking the secrets to human NTCP structure. Innovation (Camb). 2022 Aug 1;3(5):100294. DOI: 10.1016/j.xinn.2022.100294.
- ↑ 6.0 6.1 Zhang X, Zhang Q, Peng Q, Zhou J, Liao L, Sun X, Zhang L, Gong T. Hepatitis B virus preS1-derived lipopeptide functionalized liposomes for targeting of hepatic cells. Biomaterials. 2014 Jul;35(23):6130-41. DOI: 10.1016/j.biomaterials.2014.04.037.
- ↑ 7.0 7.1 7.2 Patton JS, Carey MC. Watching fat digestion. Science. 1979 Apr 13;204(4389):145-8. DOI: 10.1126/science.432636.
- ↑ Donkers JM, Kooijman S, Slijepcevic D, Kunst RF, Roscam Abbing RL, Haazen L, de Waart DR, Levels JH, Schoonjans K, Rensen PC, Oude Elferink RP, van de Graaf SF. NTCP deficiency in mice protects against obesity and hepatosteatosis. JCI Insight. 2019 Jun 25;5(14):e127197. DOI: 10.1172/jci.insight.127197.
- ↑ 9.0 9.1 Shimura S, Watashi K, Fukano K, Peel M, Sluder A, Kawai F, Iwamoto M, Tsukuda S, Takeuchi JS, Miyake T, Sugiyama M, Ogasawara Y, Park SY, Tanaka Y, Kusuhara H, Mizokami M, Sureau C, Wakita T. Cyclosporin derivatives inhibit hepatitis B virus entry without interfering with NTCP transporter activity. J Hepatol. 2017 Apr;66(4):685-692. doi: 10.1016/j.jhep.2016.11.009. DOI: 10.1016/j.jhep.2016.11.009.
Student Contributors
- Ben Minor
- Maggie Samm
- Zac Stanley