BASIL2023GV1ZBS
From Proteopedia
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
=='''''Inquiry of the Possible Function of Protein 1ZBS'''''== | =='''''Inquiry of the Possible Function of Protein 1ZBS'''''== | ||
==''' Abstract'''== | ==''' Abstract'''== | ||
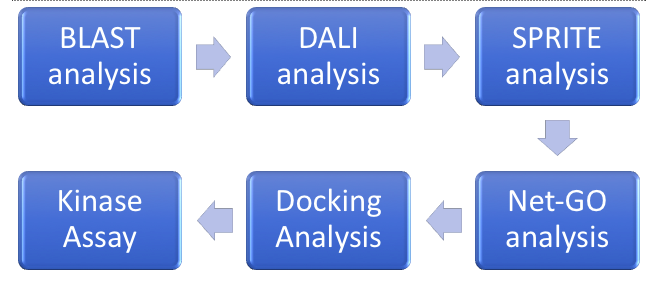
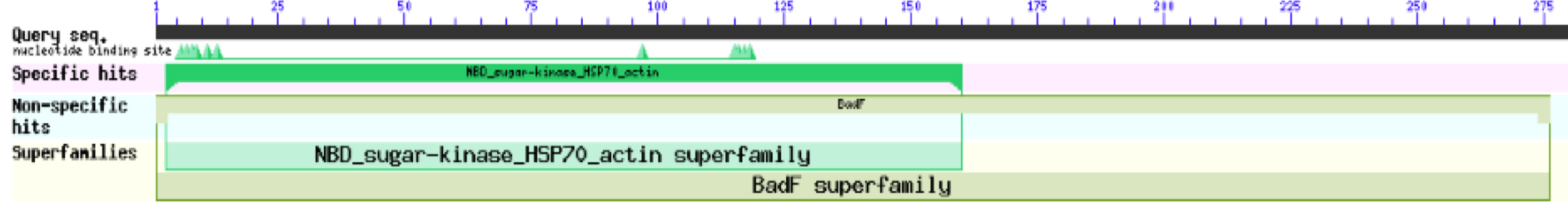
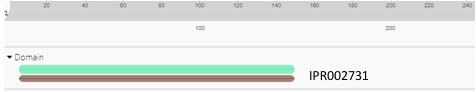
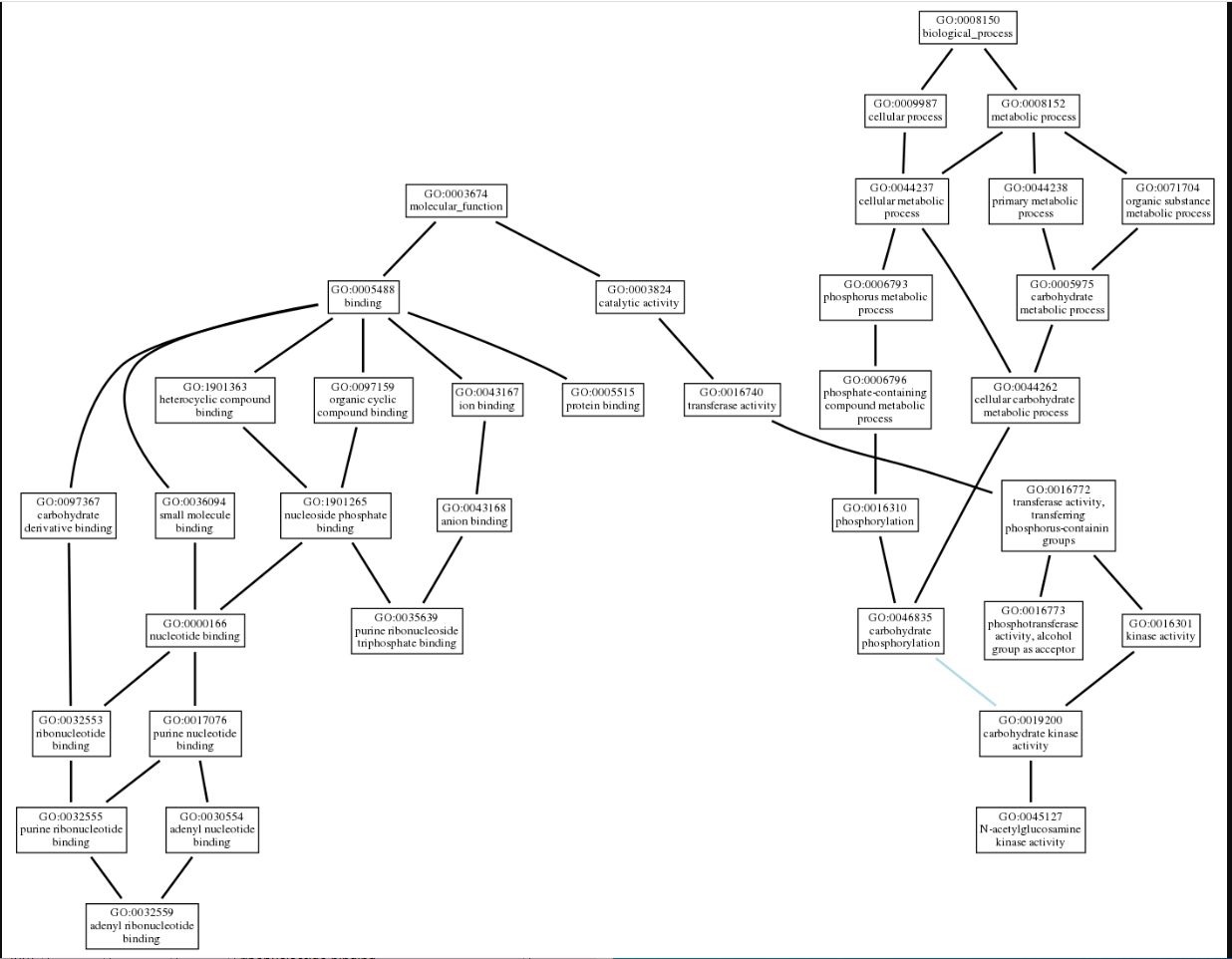
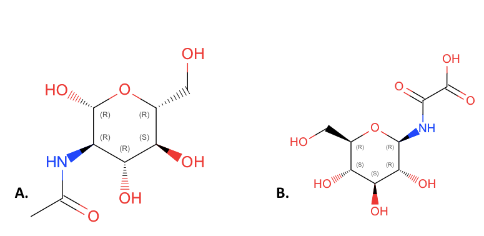
| - | ZBS is a novel protein whose structure is solved but the function is unknown. This research was designed to attempt to uncover the function. Computational research indicated that N-acetylglucosamine (NAG) may be the potential substrate, and that the protein may phosphorylate NAG. This was determined using multiple computational tools, such as BLAST-P, DALI, SPRITE, InterPro. Molecular docking using NAG as a substrate was done with PyMol and Vina docking. After the computational research was completed, the protein was over-expressed and purified. The protein was used to test for activity with the substrate NAG. The kinase assay concluded that NAG is most likely not the substrate for 1ZBS due to a lack of specific activity. | + | ZBS is a novel protein whose structure is solved but the function is unknown. This research was designed to attempt to uncover the function. Computational research indicated that N-acetylglucosamine (NAG) may be the potential substrate, and that the protein may phosphorylate NAG. This was determined using multiple computational tools, such as BLAST-P, DALI, SPRITE, InterPro. Molecular docking using NAG as a substrate was done with PyMol and Vina docking. After the computational research was completed, the protein was over-expressed and purified. The protein was used to test for activity with the substrate NAG. The kinase assay concluded that NAG is most likely not the substrate for 1ZBS due to a lack of specific activity. |
== '''Introduction''' == | == '''Introduction''' == | ||
<StructureSection load='1zbs' size='340' side='right' caption='1ZBS dimer as in its original state' scene=''> | <StructureSection load='1zbs' size='340' side='right' caption='1ZBS dimer as in its original state' scene=''> | ||
| Line 45: | Line 45: | ||
== '''Kinase Assays''' == | == '''Kinase Assays''' == | ||
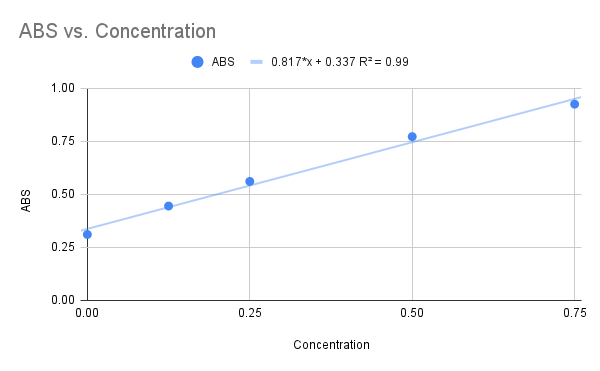
| - | We used NAG as our final substrate for the 1ZBS kinase assay, but before the kinase assay could be run, a Bradford Assay was needed to determine the concentration of the 1ZBS protein that was over-expressed and purified in the lab. The Bradford Assay gave us the graph below with an R squared value of 0.99, and the equation listed on the graph. | + | We used NAG as our final substrate for the 1ZBS kinase assay, but before the kinase assay could be run, a Bradford Assay was needed to determine the concentration of the 1ZBS protein that was over-expressed and purified in the lab. The Bradford Assay gave us the graph below with an R squared value of 0.99, and the equation listed on the graph. Because this R squared value is fairly close to 1.0, we decided to use this equation to solved for the concentration of the 1ZBS that we over-expressed. |
| + | |||
[[Image:1ZBS_Concentration.png]] | [[Image:1ZBS_Concentration.png]] | ||
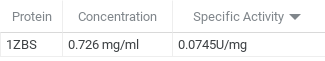
| + | After we had run the Bradford Assay, we then ran a kinase assay with 1ZBS and NAG as the substrate. From this assay, we were able to determine that the specific activity was 0.0743 U/mg which is fairly low. | ||
[[Image:1ZBS_CHART.png]] | [[Image:1ZBS_CHART.png]] | ||
== '''Conclusion''' == | == '''Conclusion''' == | ||
| Line 54: | Line 56: | ||
== References == | == References == | ||
| + | <ref>DOI:10.2210/pdb1zbs/pdb </ref> | ||
<references/> | <references/> | ||
Current revision
Inquiry of the Possible Function of Protein 1ZBS
Abstract
ZBS is a novel protein whose structure is solved but the function is unknown. This research was designed to attempt to uncover the function. Computational research indicated that N-acetylglucosamine (NAG) may be the potential substrate, and that the protein may phosphorylate NAG. This was determined using multiple computational tools, such as BLAST-P, DALI, SPRITE, InterPro. Molecular docking using NAG as a substrate was done with PyMol and Vina docking. After the computational research was completed, the protein was over-expressed and purified. The protein was used to test for activity with the substrate NAG. The kinase assay concluded that NAG is most likely not the substrate for 1ZBS due to a lack of specific activity.
Introduction
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Danielle Selover, Carmen Almendarez Rodriguez, Bonnie Hall, Jaime Prilusky