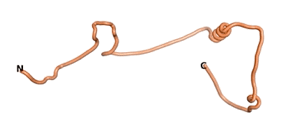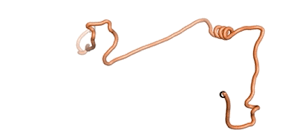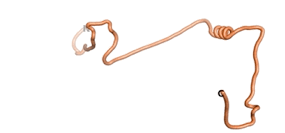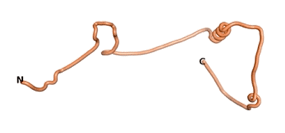Journal:Proteins:3
From Proteopedia
(Difference between revisions)

| (One intermediate revision not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size=' | + | <StructureSection load='' size='350' side='right' scene='96/964832/1hco_morph_ca/5' caption=''> |
=== Do ‘''Newly Born''’ Orphan Proteins Resemble ‘''Never Born''’ Proteins? A Study Using Three Deep Learning Algorithms === | === Do ‘''Newly Born''’ Orphan Proteins Resemble ‘''Never Born''’ Proteins? A Study Using Three Deep Learning Algorithms === | ||
<big>Jing Liu, Rongqing Yuan, Wei Shao, Jitong Wang, Israel Silman and Joel Sussman</big> <ref name="Liu">PMID:37092778</ref> | <big>Jing Liu, Rongqing Yuan, Wei Shao, Jitong Wang, Israel Silman and Joel Sussman</big> <ref name="Liu">PMID:37092778</ref> | ||
| Line 49: | Line 49: | ||
We then went on to use the three algorithms on orphan proteins and taxonomically restricted gene products (TRGP) for which no experimental structures were available. We did this in order to see how the predictions of the three algorithms would compare, and whether they would predict novel folds. Although many ORFs have been identified that code for putative orphan proteins, only in a limited number of cases has their association with a well-defined biological activity been established. We have identified seven such proteins for which the necessary sequence data are also available. The number of amino acids for these seven orphans/TRGPs ranges from 109 to 632. | We then went on to use the three algorithms on orphan proteins and taxonomically restricted gene products (TRGP) for which no experimental structures were available. We did this in order to see how the predictions of the three algorithms would compare, and whether they would predict novel folds. Although many ORFs have been identified that code for putative orphan proteins, only in a limited number of cases has their association with a well-defined biological activity been established. We have identified seven such proteins for which the necessary sequence data are also available. The number of amino acids for these seven orphans/TRGPs ranges from 109 to 632. | ||
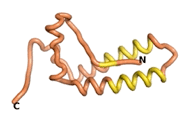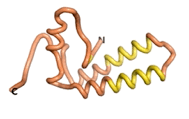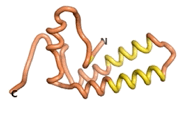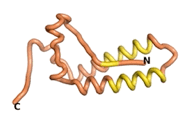
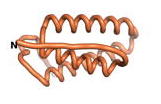
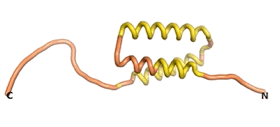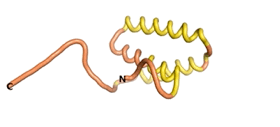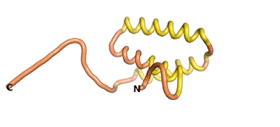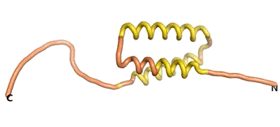
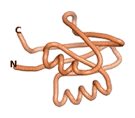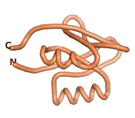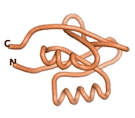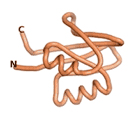
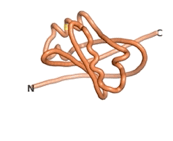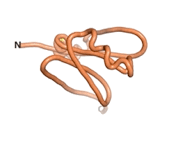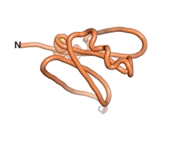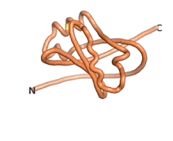
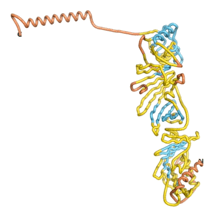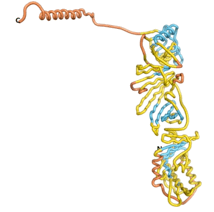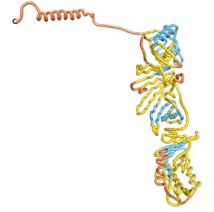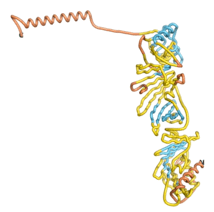
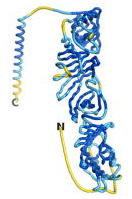
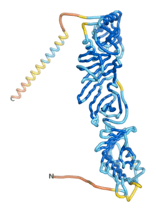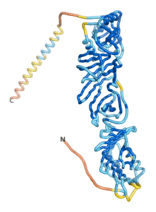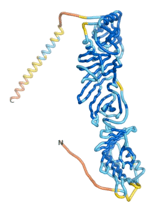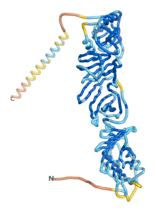
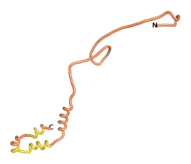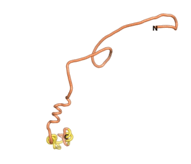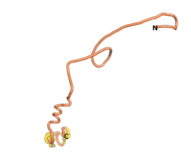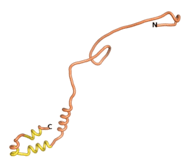
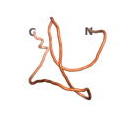
| - | As an initial step in characterizing these seven proteins, we utilized FoldIndex<ref name="FoldIndex">PMID:15955783 </ref> and flDPnn<ref name="flDPnn">PMID:34290238</ref> to investigate whether they were predicted to be intrinsically disordered proteins (IDP) or folded. Five of the proteins are predicted to be almost completely folded, while the other two, TaFROG and Newtic1, are classified as IDPs since they are predicted to be disordered throughout almost their entire sequences. Of the seven proteins studied, only HCO_011565, a 632 residue nematode protein that was shown to be the target of the nematodicidal small molecule, appears to be fully folded. The three algorithms predict almost identical structures as well as very high pLDDT scores. Most likely, this is for two reasons. Firstly, rather than being a true orphan, HCO_011565 is the product of a TRG<ref name="HCO_011565">PMID: 36313370</ref>, with the BLAST search has revealed that the first 74 homologous sequences, with the lowest E values, were all from nematodes. Secondly, the DALI server revealed a number of hits for the entire predicted structure, as well as for the three subdomains predicted by all three algorithms. It is striking just how these three different algorithms were able to predict virtually identical 3D models of this TRG, ''i.e.'', HCO_011565, with relatively high pLDDT scores for ESM and AF2, ''i.e.'', 86.2, 83.2, respectively. A 3D applet of the 5 top models of AF-2's prediction | + | As an initial step in characterizing these seven proteins, we utilized FoldIndex<ref name="FoldIndex">PMID:15955783 </ref> and flDPnn<ref name="flDPnn">PMID:34290238</ref> to investigate whether they were predicted to be intrinsically disordered proteins (IDP) or folded. Five of the proteins are predicted to be almost completely folded, while the other two, TaFROG and Newtic1, are classified as IDPs since they are predicted to be disordered throughout almost their entire sequences. Of the seven proteins studied, only HCO_011565, a 632 residue nematode protein that was shown to be the target of the nematodicidal small molecule, appears to be fully folded. The three algorithms predict almost identical structures as well as very high pLDDT scores. Most likely, this is for two reasons. Firstly, rather than being a true orphan, HCO_011565 is the product of a TRG<ref name="HCO_011565">PMID: 36313370</ref>, with the BLAST search has revealed that the first 74 homologous sequences, with the lowest E values, were all from nematodes. Secondly, the DALI server revealed a number of hits for the entire predicted structure, as well as for the three subdomains predicted by all three algorithms. It is striking just how these three different algorithms were able to predict virtually identical 3D models of this TRG, ''i.e.'', HCO_011565, with relatively high pLDDT scores for ESM and AF2, ''i.e.'', 86.2, 83.2, respectively. A 3D applet of a morph of the 5 top models of AF-2's prediction is shown to the right, and the predictions of all three algorithms are shown just below. |
{| | {| | ||
|- | |- | ||
Current revision
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.