We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1ktz
From Proteopedia
(Difference between revisions)
| (13 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | [[Image:1ktz.gif|left|200px]] | ||
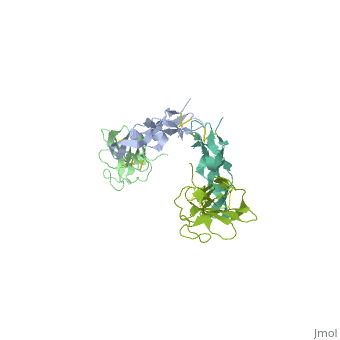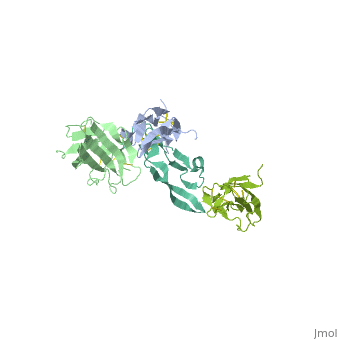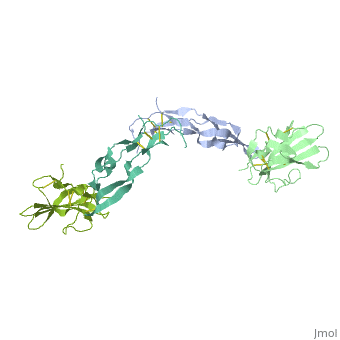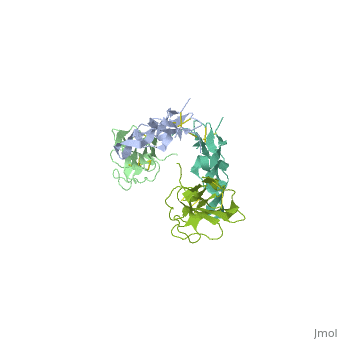
| - | + | ==Crystal Structure of the Human TGF-beta Type II Receptor Extracellular Domain in Complex with TGF-beta3== | |
| - | + | <StructureSection load='1ktz' size='340' side='right'caption='[[1ktz]], [[Resolution|resolution]] 2.15Å' scene=''> | |
| - | You may | + | == Structural highlights == |
| - | + | <table><tr><td colspan='2'>[[1ktz]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1KTZ OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1KTZ FirstGlance]. <br> | |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.15Å</td></tr> | |
| - | -- | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ktz FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ktz OCA], [https://pdbe.org/1ktz PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ktz RCSB], [https://www.ebi.ac.uk/pdbsum/1ktz PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ktz ProSAT]</span></td></tr> |
| - | + | </table> | |
| + | == Disease == | ||
| + | [https://www.uniprot.org/uniprot/TGFB3_HUMAN TGFB3_HUMAN] Defects in TGFB3 are a cause of familial arrhythmogenic right ventricular dysplasia type 1 (ARVD1) [MIM:[https://omim.org/entry/107970 107970]; also known as arrhythmogenic right ventricular cardiomyopathy 1 (ARVC1). ARVD is an autosomal dominant disease characterized by partial degeneration of the myocardium of the right ventricle, electrical instability, and sudden death. It is clinically defined by electrocardiographic and angiographic criteria; pathologic findings, replacement of ventricular myocardium with fatty and fibrous elements, preferentially involve the right ventricular free wall.<ref>PMID:15639475</ref> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/TGFB3_HUMAN TGFB3_HUMAN] Involved in embryogenesis and cell differentiation. | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/kt/1ktz_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ktz ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Transforming growth factor-beta (TGF-beta) is the prototype of a large family of structurally related cytokines that play key roles in maintaining cellular homeostasis by signaling through two classes of functionally distinct Ser/Thr kinase receptors, designated as type I and type II. TGF-beta initiates receptor assembly by binding with high affinity to the type II receptor. Here, we present the 2.15 A crystal structure of the extracellular ligand-binding domain of the human TGF-beta type II receptor (ecTbetaR2) in complex with human TGF-beta3. ecTbetaR2 interacts with homodimeric TGF-beta3 by binding identical finger segments at opposite ends of the growth factor. Relative to the canonical 'closed' conformation previously observed in ligand structures across the superfamily, ecTbetaR2-bound TGF-beta3 shows an altered arrangement of its monomeric subunits, designated the 'open' conformation. The mode of TGF-beta3 binding shown by ecTbetaR2 is compatible with both ligand conformations. This, in addition to the predicted mode for TGF-beta binding to the type I receptor ectodomain (ecTbetaR1), suggests an assembly mechanism in which ecTbetaR1 and ecTbetaR2 bind at adjacent positions on the ligand surface and directly contact each other via protein--protein interactions. | ||
| - | + | Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex.,Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP Nat Struct Biol. 2002 Mar;9(3):203-8. PMID:11850637<ref>PMID:11850637</ref> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |
| - | + | </div> | |
| + | <div class="pdbe-citations 1ktz" style="background-color:#fffaf0;"></div> | ||
| - | == | + | ==See Also== |
| - | + | *[[TGF-beta receptor|TGF-beta receptor]] | |
| + | *[[TGF-beta receptor 3D structures|TGF-beta receptor 3D structures]] | ||
| + | == References == | ||
| + | <references/> | ||
| + | __TOC__ | ||
| + | </StructureSection> | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
| - | [[Category: | + | [[Category: Large Structures]] |
| - | + | [[Category: Deep S]] | |
| - | [[Category: Deep | + | [[Category: Hart PJ]] |
| - | [[Category: Hart | + | [[Category: Hinck AP]] |
| - | [[Category: Hinck | + | [[Category: Hinck CS]] |
| - | [[Category: Hinck | + | [[Category: Shu Z]] |
| - | [[Category: Shu | + | [[Category: Taylor AB]] |
| - | [[Category: Taylor | + | |
| - | + | ||
| - | + | ||
Current revision
Crystal Structure of the Human TGF-beta Type II Receptor Extracellular Domain in Complex with TGF-beta3
| |||||||||||
Categories: Homo sapiens | Large Structures | Deep S | Hart PJ | Hinck AP | Hinck CS | Shu Z | Taylor AB