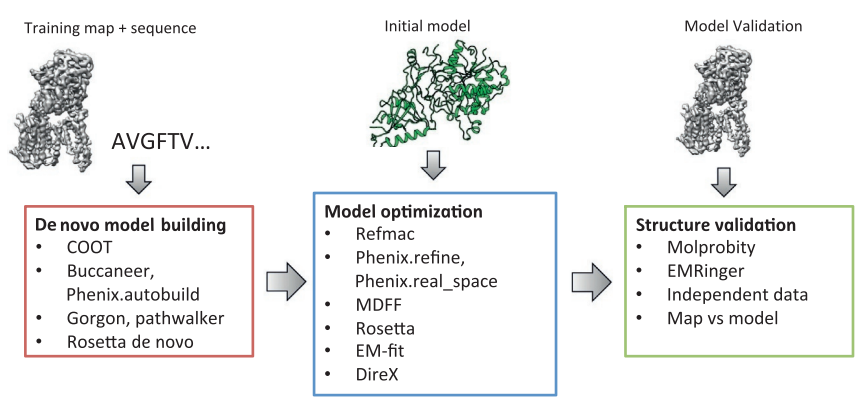
User:Francielle Aguiar Gomes/Sandbox 1
From Proteopedia
< User:Francielle Aguiar Gomes(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 16: | Line 16: | ||
[[Image:Membrane.png]] | [[Image:Membrane.png]] | ||
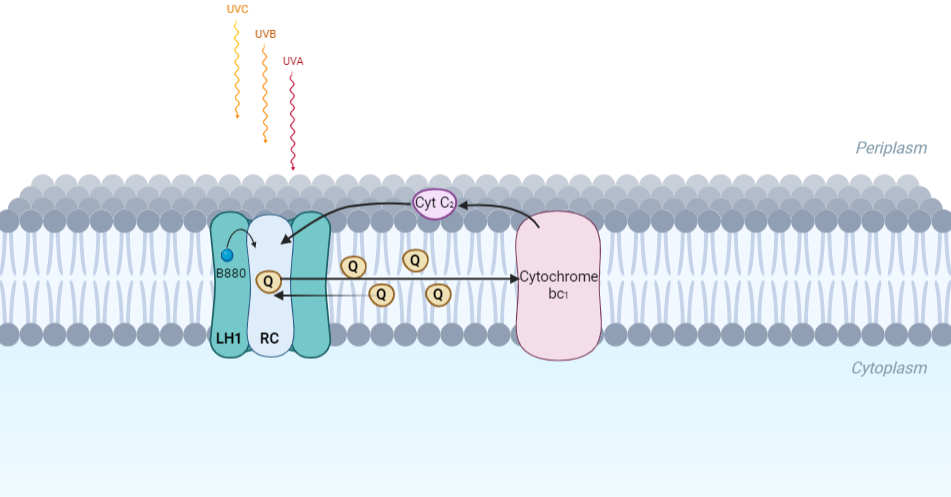
| - | Representation of the cyclic electronic transport process in the intracytoplasmic membrane of purple bacteria. Light is absorbed by the LHC1 complex, which transfer their excitation energy to the reaction center, where a separation of loads. The light energy absorbed by carotenoids and bacteriochlorophylls ( | + | Representation of the cyclic electronic transport process in the intracytoplasmic membrane of purple bacteria. Light is absorbed by the LHC1 complex, which transfer their excitation energy to the reaction center, where a separation of loads. The light energy absorbed by carotenoids and bacteriochlorophylls (B820) generates a change in the energy state of the molecules that can be transferred following several excitation pathways between the photosystem pigments until it ends up reducing the ubiquinones located in the RC (thus converting light energy into chemical energy). They escape through interprotein pores in the RC to transfer electrons to Cyt b1. The route is completed by a soluble protein (Cyt c2) that ends up donating electrons to LH1. |
== Inicial Structures == | == Inicial Structures == | ||
| Line 43: | Line 43: | ||
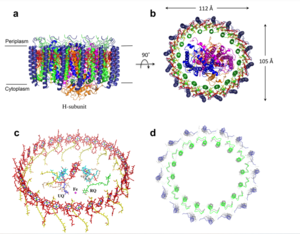
For a more comprehensive view of the structure of ''Rsp. rubrum'' LH1-RC, this molecule can be visualized in different ways, such as, for instance, by the shape of <scene name='96/969634/Backbone/1'>backbone</scene>, <scene name='96/969634/Ballandstick/1'>Ball and Stick</scene> or <scene name='96/969634/Spacefill/1'>spacefill</scene>. | For a more comprehensive view of the structure of ''Rsp. rubrum'' LH1-RC, this molecule can be visualized in different ways, such as, for instance, by the shape of <scene name='96/969634/Backbone/1'>backbone</scene>, <scene name='96/969634/Ballandstick/1'>Ball and Stick</scene> or <scene name='96/969634/Spacefill/1'>spacefill</scene>. | ||
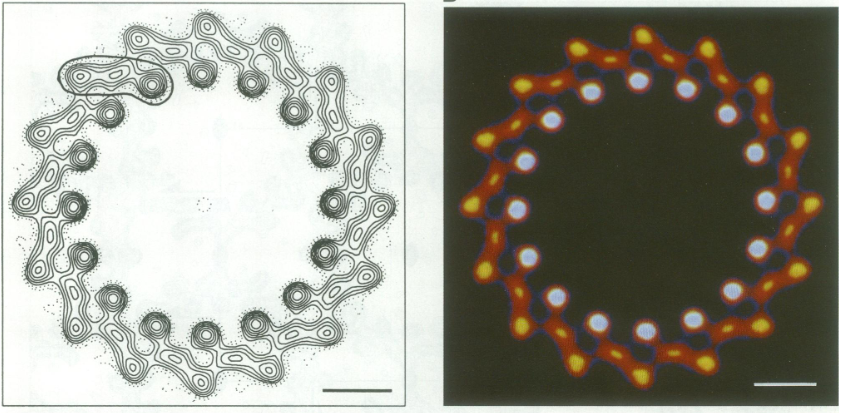
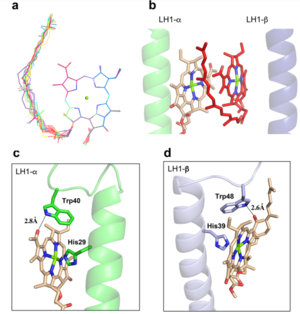
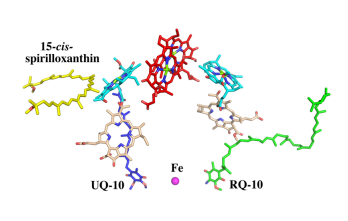
| - | The BChl aG molecules in ''Rsp. rubrum'' LH1 forms an elliptical, partially overlapping ring with average Mg−Mg distances of 9.3 Å within a dimer and 8.5 Å between dimers (Figure 1c). These molecules are ligated by histidine residues (α-His29 and β-His39), as shown in the next image. The geranylgeranyl side chains in the BChl aG associated with βpolypeptides form a tail-up conformation (Figure 2a). This may be due to the multiple double bonds present in the geranylgeranyl group, which results in a more rigid conformation. This unique conformation allows the geranylgeranyl side chains of the β-associated BChl aG to interact with the bacteriochlorin ring of the α-associated BChl aG (Figure | + | The BChl aG molecules in ''Rsp. rubrum'' LH1 forms an elliptical, partially overlapping ring with average Mg−Mg distances of 9.3 Å within a dimer and 8.5 Å between dimers (Figure 1c). These molecules are ligated by histidine residues (α-His29 and β-His39), as shown in the next image. The geranylgeranyl side chains in the BChl aG associated with βpolypeptides form a tail-up conformation (Figure 2a). This may be due to the multiple double bonds present in the geranylgeranyl group, which results in a more rigid conformation. This unique conformation allows the geranylgeranyl side chains of the β-associated BChl aG to interact with the bacteriochlorin ring of the α-associated BChl aG (Figure 2b). This proximity likely allows for π−π interactions between the double bonds in the geranylgeranyl group and nearby bacteriochlorins. The C3-acetyl oxygen atoms in BChl aG form hydrogen bonds with the Trp residues (α-Trp40 and β-Trp48) of their associated polypeptides (Figure 2c, d), in agreement with the results of resonance Raman spectroscopy. The aromatic side chains of the Trp residues also interact with bacteriochlorins through π−π stacking, further stabilizing the LH1 complex. <ref>10.1073/pnas.91.15.7124</ref><ref>10.1016/0005-2728(85)90048-9</ref> |
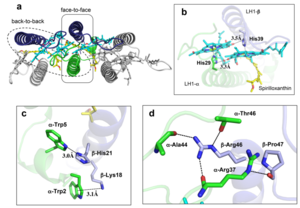
[[Image:Ligant.png|300px|right|thumb| '''Fig. 2.''' (a) Superposition of the bacteriochlorin rings of 16 BChl aG molecules bound to the LH1 βpolypeptides. (b) Interactions between a geranylgeranyl side chain of the BChl aG (red sticks) bound to LH1 β-polypeptide and the bacteriochlorin ring (tint sticks) of a BChl aG bound to the LH1 α-polypeptide. (c) Coordination and hydrogen bonding of the BChl aG in LH1 α-polypeptide. (d) Coordination and hydrogen bonding of the BChl aG in LH1 β-polypeptide.]] | [[Image:Ligant.png|300px|right|thumb| '''Fig. 2.''' (a) Superposition of the bacteriochlorin rings of 16 BChl aG molecules bound to the LH1 βpolypeptides. (b) Interactions between a geranylgeranyl side chain of the BChl aG (red sticks) bound to LH1 β-polypeptide and the bacteriochlorin ring (tint sticks) of a BChl aG bound to the LH1 α-polypeptide. (c) Coordination and hydrogen bonding of the BChl aG in LH1 α-polypeptide. (d) Coordination and hydrogen bonding of the BChl aG in LH1 β-polypeptide.]] | ||
| Line 57: | Line 57: | ||
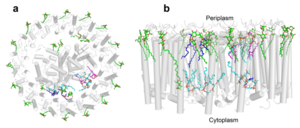
[[Image:Peri.png|300px|left|thumb| '''Fig. 3.''' Phospholipids, detergents, and channels in the LH1-RC complex. Top view (a) and side view (b) of the phospholipid and detergent distributions for CL (cyan), PG (magenta), PE (blue), and DDM (green). All proteins are shown in gray.]] | [[Image:Peri.png|300px|left|thumb| '''Fig. 3.''' Phospholipids, detergents, and channels in the LH1-RC complex. Top view (a) and side view (b) of the phospholipid and detergent distributions for CL (cyan), PG (magenta), PE (blue), and DDM (green). All proteins are shown in gray.]] | ||
| - | The ''Rsp. rubrum'' LH1 complex is particularly well-known for its ability to form a highly stable structural subunit with an absorption maximum of | + | The ''Rsp. rubrum'' LH1 complex is particularly well-known for its ability to form a highly stable structural subunit with an absorption maximum of 875 nm. Two different subunit forms can be distinguished from the LH1 complex: a face-to-face and a back-to-back configuration for the bacteriochlorins. Solution NMR and reconstitution experiments established that the B820 subunit has the face-to-face configuration with π-overlap at pyrrole rings III and V.<ref>10.1042/BCJ20160753</ref> |
Due to its structural simplicity and flexibility, the ''Rsp. rubrum'' B820 subunit has been thoroughly investigated and the results from which predicted minimal requirements for stabilizing the subunit structure. These requirements included (i) a central α-helical transmembrane domain composed of 18 hydrophobic residues, (ii) a His residue for coordination and hydrogen bonding to different BChl molecules, (iii) a Trp residue for hydrogen bonding to the BChl C31 carbonyl oxygen, and (iv) N-terminal regions of α- and βpolypeptides. The His interaction was estimated to account for over half of the stabilization energy of the B820 subunit, followed by hydrogen bonding by Trp residues.<ref>10.1021/bi9722709</ref><ref>10.1039/C7SC04905F</ref> | Due to its structural simplicity and flexibility, the ''Rsp. rubrum'' B820 subunit has been thoroughly investigated and the results from which predicted minimal requirements for stabilizing the subunit structure. These requirements included (i) a central α-helical transmembrane domain composed of 18 hydrophobic residues, (ii) a His residue for coordination and hydrogen bonding to different BChl molecules, (iii) a Trp residue for hydrogen bonding to the BChl C31 carbonyl oxygen, and (iv) N-terminal regions of α- and βpolypeptides. The His interaction was estimated to account for over half of the stabilization energy of the B820 subunit, followed by hydrogen bonding by Trp residues.<ref>10.1021/bi9722709</ref><ref>10.1039/C7SC04905F</ref> | ||
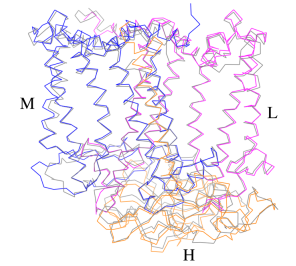
Additionally, our ''Rsp. rubrum'' LH1 structure also underscores the importance of the C-terminal domains (Figure 4d), especially for the β-polypeptide where the amino acids are highly conserved. The side chain of β-Arg46 forms multiple hydrogen bonds with the hydroxyl group of α-Thr46 and main chain oxygen atoms of α-Arg37 and α-Ala44. β-Arg46 is conserved in the LH1 of almost all purple bacteria and contributes 2.0 kcal/mol stabilization energy to the B820 subunit and this follows only the pigment−protein interactions of BChl a/β-His39 (>6 kcal/mol) and BChl a/β-Trp48 (3.7 kcal/mol) in stabilization energy.<ref>10.1023/A:1006337827672</ref><ref>10.1021/bi049798f</ref> | Additionally, our ''Rsp. rubrum'' LH1 structure also underscores the importance of the C-terminal domains (Figure 4d), especially for the β-polypeptide where the amino acids are highly conserved. The side chain of β-Arg46 forms multiple hydrogen bonds with the hydroxyl group of α-Thr46 and main chain oxygen atoms of α-Arg37 and α-Ala44. β-Arg46 is conserved in the LH1 of almost all purple bacteria and contributes 2.0 kcal/mol stabilization energy to the B820 subunit and this follows only the pigment−protein interactions of BChl a/β-His39 (>6 kcal/mol) and BChl a/β-Trp48 (3.7 kcal/mol) in stabilization energy.<ref>10.1023/A:1006337827672</ref><ref>10.1021/bi049798f</ref> | ||
Current revision
Photosynthetic LH1-RC Super-complex of Rhodospirillum rubrum
| |||||||||||