User:Marcos Vinícius Caetano/Sandbox 1
From Proteopedia
< User:Marcos Vinícius Caetano(Difference between revisions)
| (4 intermediate revisions not shown.) | |||
| Line 24: | Line 24: | ||
== ATPase cycle of myosin VI== | == ATPase cycle of myosin VI== | ||
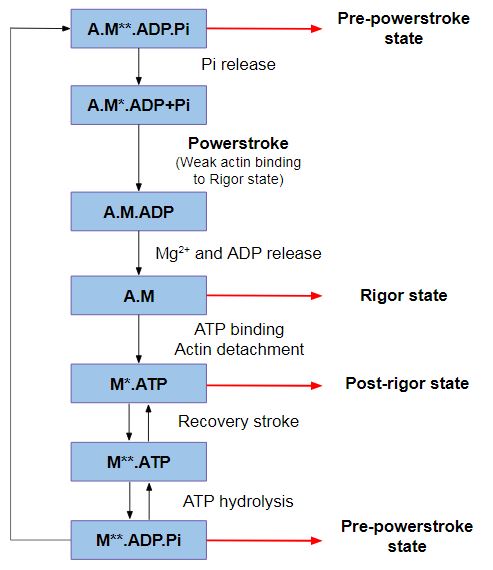
| - | Despite the fact that myosin VI directionality is reversed, it has the '''same kinetic ATPase cycle''' of interaction with actin, as shown in the figure below. There are 3 states of different conformation: Pre-powerstroke, Rigor state and Post-rigor state <ref>Ménétrey, J., Isabet, T., Ropars, V., Mukherjea, M., Pylypenko, O., Liu, X., Perez, J., Vachette, P., Sweeney, H. L., & Houdusse, A. M. (2012). Processive steps in the reverse direction require uncoupling of the lead head lever arm of myosin VI. Molecular cell, 48(1), 75–86. https://doi.org/10.1016/j.molcel.2012.07.034</ref>. | + | Despite the fact that myosin VI directionality is reversed, it has the '''same kinetic ATPase cycle''' of interaction with actin, as shown in the figure below. There are 3 states of different conformation: Pre-powerstroke, Rigor state and Post-rigor state <ref>Ménétrey, J., Isabet, T., Ropars, V., Mukherjea, M., Pylypenko, O., Liu, X., Perez, J., Vachette, P., Sweeney, H. L., & Houdusse, A. M. (2012). Processive steps in the reverse direction require uncoupling of the lead head lever arm of myosin VI. Molecular cell, 48(1), 75–86. https://doi.org/10.1016/j.molcel.2012.07.034</ref>. <ref>Ménétrey, J., Llinas, P., Cicolari, J., Squires, G., Liu, X., Li, A., Sweeney, H. L., & Houdusse, A. (2008). The post-rigor structure of myosin VI and implications for the recovery stroke. The EMBO journal, 27(1), 244–252. https://doi.org/10.1038/sj.emboj.7601937</ref> |
[[Image:Cycle ATP.JPG|500px]] | [[Image:Cycle ATP.JPG|500px]] | ||
''Figure 2: ATPase cycle of myosins.'' | ''Figure 2: ATPase cycle of myosins.'' | ||
| + | |||
'''Pre-powerstroke''': myosin binds to actin, it has hydrolyzed ATP, but keeps the products MgADP and Pi. The interaction of myosin with actin results in the release of Pi, which increases the binding affinity for actin and triggers the release of MgADP to form a rigor conformation on actin. At this moment, occurs the powerstroke, a large movement (11 nm) of the lever arm between the pre-powerstroke and rigor state, during which the converter and the CaM rotates and alters its conformation. | '''Pre-powerstroke''': myosin binds to actin, it has hydrolyzed ATP, but keeps the products MgADP and Pi. The interaction of myosin with actin results in the release of Pi, which increases the binding affinity for actin and triggers the release of MgADP to form a rigor conformation on actin. At this moment, occurs the powerstroke, a large movement (11 nm) of the lever arm between the pre-powerstroke and rigor state, during which the converter and the CaM rotates and alters its conformation. | ||
| Line 54: | Line 55: | ||
*'''Insert 2''' (<scene name='97/973101/774-812_orange/2'>Pro774-Tyr812</scene>) - it redirects the lever arm towards the minus end. It also contains a new calmodulin-binding motif. | *'''Insert 2''' (<scene name='97/973101/774-812_orange/2'>Pro774-Tyr812</scene>) - it redirects the lever arm towards the minus end. It also contains a new calmodulin-binding motif. | ||
*'''<scene name='97/973101/Iq_motif/1'>IQ motif</scene>''' - Constitute the binding sites for Calcium-free, Calcium-calmodulin (CaM) or Apocalmodulin (apo-CaM). | *'''<scene name='97/973101/Iq_motif/1'>IQ motif</scene>''' - Constitute the binding sites for Calcium-free, Calcium-calmodulin (CaM) or Apocalmodulin (apo-CaM). | ||
| - | *'''<scene name='97/973101/P_loop_switch_i_and_ii/1'>P-loop, switch I and switch II</scene>''': <span style="color:blue">'''P-loop'''</span>, <span style="color:magenta">'''Switch I'''</span> and <span style="color:green">'''Switch II'''</span> are nucleotide-binding elements, participating in nucleotide-binding in different stages of the kinetic cycle. More specifically, | + | *'''<scene name='97/973101/P_loop_switch_i_and_ii/1'>P-loop, switch I and switch II</scene>''': <span style="color:blue">'''P-loop'''</span>, <span style="color:magenta">'''Switch I'''</span> and <span style="color:green">'''Switch II'''</span> are nucleotide-binding elements, participating in nucleotide-binding in different stages of the kinetic cycle. More specifically, <span style="color:magenta">'''Switch I'''</span> binds both the Mg<sup>2+</sup> ion and the γ-phosphate of ATP in the active site, therefore, it controls the nucleotide release from the motor and binding to the motor. The <scene name='97/973101/P_loop_switch_i_and_ii/2'>spacefill representation</scene> shows a better view where the nucleotides binds and how tightly this process may be regulated. |
| - | The '''motor domain''' contains '''two inserts''' that are unique in the myosin superfamily: | + | The '''motor domain''' of myosin VI contains '''two inserts''' that are unique in the myosin superfamily: |
'''Insert 1: <scene name='97/973101/278_303_purple/1'>Cys278-Ala303</scene>.''' | '''Insert 1: <scene name='97/973101/278_303_purple/1'>Cys278-Ala303</scene>.''' | ||
| Line 65: | Line 66: | ||
*'''Function''': This insert provides unique kinetic characteristics: it modulates nucleotide binding and Switch I flexibility, therefore, it slows ADP release and ATP-induced dissociation of the motor from actin (at saturating ATP concentrations). | *'''Function''': This insert provides unique kinetic characteristics: it modulates nucleotide binding and Switch I flexibility, therefore, it slows ADP release and ATP-induced dissociation of the motor from actin (at saturating ATP concentrations). | ||
| - | *'''Mechanism''': As also seen in all other myosins, the conformation of Switch I relative to the U50kDa subdomain is not altered by the presence of insert 1. However, the small loop (<scene name='97/973101/Insert_1_and_small_loop/1'>Gly304-Asp313</scene> - in <span style="color:grey">'''grey'''</span>) that follows this insert, is repositioned, standing out in the nucleotide-binding pocket (decreasing nucleotide accessibility by steric impediment) and strongly interacting with Switch I by the residues: <scene name='97/973101/Insert_1_and_small_loop_key/1'>L306, D308, L310, L311</scene> (in <span style="color:red">'''red'''</span>). Also, <scene name='97/973101/Insert_1_and_small_loop_key/1'>C278 and F282</scene> (in <span style="color:green">'''green'''</span>) of insert 1 interacts with <scene name='97/973101/Insert_1_and_small_loop_key/3'>Switch I</scene> (in <span style="color:magenta">'''magenta'''</span>). <scene name='97/973101/Insert_1_and_small_loop_key/2'>Leucine 310</scene> (highlighted in <span style="color:blue">'''blue'''</span>) is specifically important because its position selectively interferes with ATP binding, while having little or no effect on ADP binding. Mutation of | + | *'''Mechanism''': As also seen in all other myosins, the conformation of Switch I relative to the U50kDa subdomain is not altered by the presence of insert 1. However, the small loop (<scene name='97/973101/Insert_1_and_small_loop/1'>Gly304-Asp313</scene> - in <span style="color:grey">'''grey'''</span>) that follows this insert, is repositioned, standing out in the nucleotide-binding pocket (decreasing nucleotide accessibility by steric impediment) and strongly interacting with Switch I by the residues: <scene name='97/973101/Insert_1_and_small_loop_key/1'>L306, D308, L310, L311</scene> (in <span style="color:red">'''red'''</span>). Also, <scene name='97/973101/Insert_1_and_small_loop_key/1'>C278 and F282</scene> (in <span style="color:green">'''green'''</span>) of insert 1 interacts with <scene name='97/973101/Insert_1_and_small_loop_key/3'>Switch I</scene> (in <span style="color:magenta">'''magenta'''</span>). <scene name='97/973101/Insert_1_and_small_loop_key/2'>Leucine 310</scene> (highlighted in <span style="color:blue">'''blue'''</span>) is specifically important because its position selectively interferes with ATP binding, while having little or no effect on ADP binding. Mutation of Leucine 310 to Glycine removes all influence of insert 1 on ATP binding <ref>Pylypenko, O., Song, L., Squires, G., Liu, X., Zong, A. B., Houdusse, A., & Sweeney, H. L. (2011). Role of insert-1 of myosin VI in modulating nucleotide affinity. The Journal of biological chemistry, 286(13), 11716–11723. https://doi.org/10.1074/jbc.M110.200626</ref>. |
| Line 75: | Line 76: | ||
*'''Function''': Redirectionare the lever arm and contains a new CaM-binding motif. | *'''Function''': Redirectionare the lever arm and contains a new CaM-binding motif. | ||
| - | *'''Mechanism''': The proximal part of insert 2 (<scene name='97/973101/774_812_distal_and_proximal/3'>Pro774-Trp787</scene> - in <span style="color:red">'''red'''</span>) wraps around the <scene name='97/973101/774_812_distal_and_proximal/4'>converter</scene> (in <span style="color:blue">'''blue'''</span>), while the distal part (<scene name='97/973101/774_812_distal_and_proximal/3'>Trp787-Tyr812</scene> - in <span style="color:green">'''green'''</span>) forms a CaM-binding motif. The insert 2 and its associated CaM molecule (with 4Ca<sup>2+</sup>), make specific interactions with the converter, many involving a variable loop (<scene name='97/973101/Insert_2_full/1'>Lys719-Pro731</scene>- in <span style="color:magenta">'''magenta'''</span>). In addiction, there is '''apolar interactions''' that stabilize the proximal part of insert 2 on the surface of the converter, where <scene name='97/973101/Insert_2_full/3'>hidrophofobic side chains</scene> from the amino acids <span style="color:gold">'''F763, F766, M770'''</span> stabilize the orientation of the last helix <scene name='97/973101/Insert_2_full/4'>(representation with cartoon and stick)</scene>. Also, there is <scene name='97/973101/Insert_2_full_salt_bridges/2'>salt-bridge interactions</scene> between the converter and both lobes of CaM, throghout the amino acids: <span style="color:orange">''' D730, E114, K795, R792'''</span>; <span style="color:blue">'''K736, E14, R732'''</span>; and <span style="color:gold">'''R728, E120'''</span>. The result of <scene name='97/973101/Insert_2_full/2'>interactions</scene> is that <scene name='97/973101/Iq_helix_emerge/1'>IQ helix</scene> | + | *'''Mechanism''': The proximal part of insert 2 (<scene name='97/973101/774_812_distal_and_proximal/3'>Pro774-Trp787</scene> - in <span style="color:red">'''red'''</span>) wraps around the <scene name='97/973101/774_812_distal_and_proximal/4'>converter</scene> (in <span style="color:blue">'''blue'''</span>), while the distal part (<scene name='97/973101/774_812_distal_and_proximal/3'>Trp787-Tyr812</scene> - in <span style="color:green">'''green'''</span>) forms a CaM-binding motif. The insert 2 and its associated CaM molecule (with 4Ca<sup>2+</sup>), make specific interactions with the converter, many involving a variable loop (<scene name='97/973101/Insert_2_full/1'>Lys719-Pro731</scene>- in <span style="color:magenta">'''magenta'''</span>). In addiction, there is '''apolar interactions''' that stabilize the proximal part of insert 2 on the surface of the converter, where <scene name='97/973101/Insert_2_full/3'>hidrophofobic side chains</scene> from the amino acids <span style="color:gold">'''F763, F766, M770'''</span> stabilize the orientation of the last helix <scene name='97/973101/Insert_2_full/4'>(representation with cartoon and stick)</scene>. Also, there is <scene name='97/973101/Insert_2_full_salt_bridges/2'>salt-bridge interactions</scene> between the converter and both lobes of CaM, throghout the amino acids: <span style="color:orange">''' D730, E114, K795, R792'''</span>; <span style="color:blue">'''K736, E14, R732'''</span>; and <span style="color:gold">'''R728, E120'''</span>. The result of <scene name='97/973101/Insert_2_full/2'>interactions</scene> is that <scene name='97/973101/Iq_helix_emerge/1'>IQ helix</scene> (in <span style="color:green">'''green'''</span>) '''emerges ~120°''' from the position that it emerges in all other myosins, '''redirecting the IQ helix''' and the CaM towards the '''minus-end''' of the actin filament. '''This is what makes myosin VI unique'''. |
The figure below compare the orientation of the last helix of the converter in myosin VI (<span style="color:green">'''green'''</span>) with myosin V (<span style="color:blue">'''blue'''</span>), and it shows a '''difference of 19°''', that combined with the apolar and salt-bridges interactions, makes the emerge of the IQ helix 120° from the position that it emerges in all other myosins. | The figure below compare the orientation of the last helix of the converter in myosin VI (<span style="color:green">'''green'''</span>) with myosin V (<span style="color:blue">'''blue'''</span>), and it shows a '''difference of 19°''', that combined with the apolar and salt-bridges interactions, makes the emerge of the IQ helix 120° from the position that it emerges in all other myosins. | ||
| Line 87: | Line 88: | ||
== References == | == References == | ||
<references /> | <references /> | ||
| - | |||
| - | |||
| - | *Ménétrey, J., Llinas, P., Cicolari, J., Squires, G., Liu, X., Li, A., Sweeney, H. L., & Houdusse, A. (2008). The post-rigor structure of myosin VI and implications for the recovery stroke. The EMBO journal, 27(1), 244–252. https://doi.org/10.1038/sj.emboj.7601937 | ||
Current revision
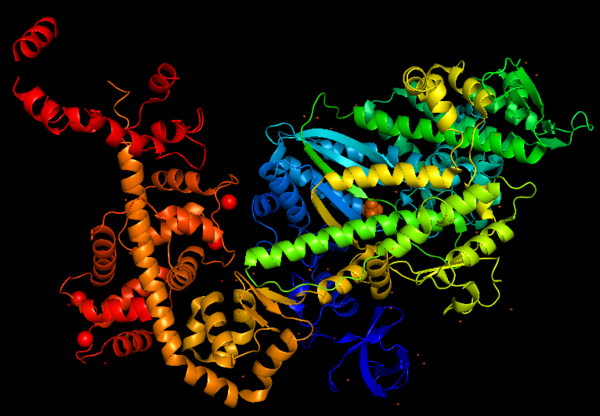
Myosin VI nucleotide-free (MDinsert2-IQ) crystal structure (2BKI)
| |||||||||||