Journal:Acta Cryst D:S2059798323006642
From Proteopedia
(Difference between revisions)

(New page: :) |
|||
| (63 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | : | + | <StructureSection load='' size='450' side='right' scene='98/982642/Cv/3' caption=''> |
| + | ===Atypical Homodimerisation Revealed by the Structure of (S)-Enantioselective Haloalkane Dehalogenase DmmarA from ''Mycobacterium marinum''=== | ||
| + | <big>Karolina Snajdarova, Sérgio M. Marques, Jiri Damborsky, David Bednar, Martin Marek</big> <ref>doi: 10.1107/S2059798323006642</ref> | ||
| + | <hr/> | ||
| + | <b>Molecular Tour</b><br> | ||
| + | Haloalkane dehalogenases are a family of alpha/beta-hydrolase fold enzymes employing S<sub>N</sub>2 nucleophilic substitution to cleave the carbon-halogen bond in diverse chemical structures, whose biological role is still poorly understood. | ||
| + | |||
| + | Haloalkane dehalogenases: | ||
| + | *DpaA - ''Paraglaciecola agarilytica'' NO2 [[7avr]] tetramer | ||
| + | *DppA - ''Plesiocystis pacifica'' SIR-1 [[2xt0]] dimer | ||
| + | *DatA - ''Agrobacterium fabrum'' C58 [[3wi7]] dimer | ||
| + | *DbeA - ''Bradyrhizobium elkanii'' USDA 94 [[4k2a]] dimer | ||
| + | *DmmA - unidentified marine microbiome [[3u1t]] dimer | ||
| + | *DmxA - ''Marinobacter sp.'' ELB17 [[5mxp]] dimer | ||
| + | *DccA - ''Caulobacter vibrioides'' CB15 [[5esr]] dimer | ||
| + | *HanR - ''Rhodobacteraceae bacterium'' UDC319 [[4brz]] dimer | ||
| + | *DbjA - ''Bradyrhizobium diazoeficiens'' USDA 110 [[3afi]] dimer | ||
| + | *LinB - ''Sphingomonas Paucimobilis'' UT26 [[1mj5]] monomer | ||
| + | |||
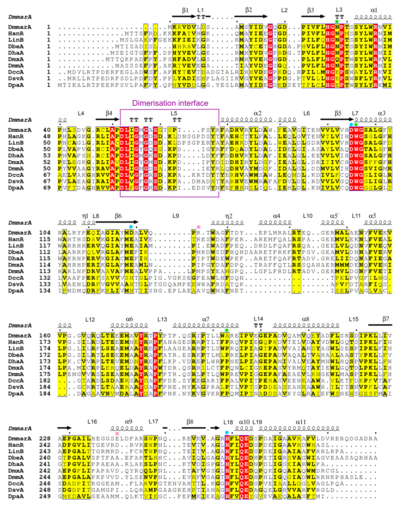
| + | [[Image:Figure3snaj1.png|left|thumb|400px|Multiple sequence alignment of different HLDs. The alignment includes sequences of DmmarA, HanR, LinB, DbeA, DhaA, DmxA, DmmA, DccA, DsvA and DpaA, and secondary structure topology of DmmarA above the aligned sequences. Residues of the catalytic triad are marked by a blue dot, halide-stabilizing residues as a green star, and atypical residues present in the catalytic cavity as a pink square. The dimerization interface is framed in purple.]] | ||
| + | {{Clear}} | ||
| + | |||
| + | Atomic-level knowledge of both the inner organization and supramolecular complexation of HLDs is thus crucial for our understanding of their catalytic and non-catalytic functions. Here, we determined crystallographic structures of an (''S'')-enantioselective haloalkane dehalogenase '''DmmarA''' from the waterborne pathogenic microbe ''Mycobacterium marinum'' at 1.6 Å and 1.85 Å resolutions. | ||
| + | |||
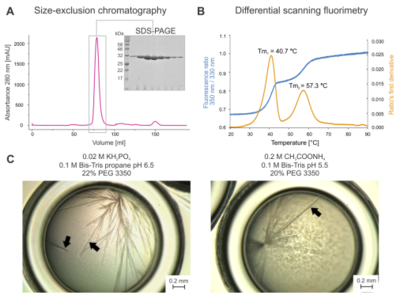
| + | [[Image:Figure1snaj1.png|left|thumb|400px|Biochemical characterisation and crystal morphology. (A) Elution profile from size-exclusion chromatography with SDS-PAGE. (B) Thermal unfolding experiments. The blue curve represents the tryptophan fluorescence ratio at 350 nm/330 nm, and the orange curve is the ratio’s first derivative. Note that two major melting points accompany the melting of DmmarA: ''T''<sub>m1</sub> = 40.7°C and ''T''<sub>m2</sub> = 57.3°C. (C) Micrographs of needle-like DmmarA crystals were obtained at pH 6.5 (left panel) and 5.5 (right panel). Black arrowheads point to diffraction-quality crystals that display three-dimensional morphology. Bars represent 0.2 mm.]] | ||
| + | {{Clear}} | ||
| + | |||
| + | Overall structure of DmmarA: | ||
| + | *<scene name='98/982642/Frontview/2'>Front view</scene>. <span class="bg-yellow"><span class="far fa-hand-point-right"></span> Remember to drag the structures with the mouse to rotate them.</span> | ||
| + | *<scene name='98/982642/Sideview/3'>Side view</scene>. | ||
| + | *<scene name='98/982642/Bottom_view/1'>Bottom view</scene>. | ||
| + | Chain A is displayed as a blue cartoon, and chain B as a light blue cartoon. Amino acids forming the catalytic pentad are visualized as green spheres. The L5 loop is colored purple. | ||
| + | |||
| + | The structures show a canonical αβα-sandwich HLD fold with several unusual structural features. Mechanistically, the atypical composition of a proton-relay catalytic triad (aspartate-histidine-aspartate) and uncommon active site pocket expose molecular specificities of catalytic apparatus that exhibits rare (''S'')-enantiopreference. | ||
| + | |||
| + | Active sites of DmmarA and ''Rhodobacteraceae bacterium'' UDC319 Haloalkane dehalogenase (HanR; [[4brz]]): | ||
| + | *<scene name='98/982642/Active_site/9'>Active site of DmmarA</scene>. Residues of the proton-relay system, halide-binding site and access tunnel are displayed as blue ball-and-sticks. Unique residues are colored in lavender. Hydrogen bonds between residues and with water are shown as white dashed lines. Water is visualized as a red sphere. | ||
| + | *<scene name='98/982642/Active_site/6'>The superposition of active sites of DmmarA (blue) and HanR (gold)</scene>. | ||
| + | |||
| + | Additionally, the structures reveal an as-yet-unseen mode of symmetric homodimerization, which is predominantly mediated through unusual L5-to-L5 loops interactions. <scene name='98/982642/Dimerization/1'>Back-to-back dimerization interface of DmmarA through L5 loop</scene> (chain A is displayed as a blue cartoon, and chain B as a light blue cartoon; the L5 loop is colored purple). In the <scene name='98/982642/Dimerization/4'>close-up view</scene>, interacting residues are shown as ball-and-sticks. Water molecules are shown as red spheres, bonds as white dashed lines. This homodimeric association in a solution is confirmed experimentally by data obtained from small-angle X-ray scattering. Utilizing the newly determined structures of DmmarA, molecular modelling techniques were employed to elucidate the underlying mechanism behind its uncommon enantioselectivity. The (''S'')-preference can be attributed to the presence of a distinct binding pocket and variances in the activation barrier of nucleophilic substitution. | ||
| + | |||
| + | <scene name='98/982642/Dmmara_l5_loop/3'>Unique DmmarA L5 loop</scene>. <span class="bg-yellow"><span class="far fa-hand-point-right"></span> Remember to drag the structures with the mouse to rotate them.</span> <scene name='98/982642/Dmmara_l5_loop/4'>Unique DmmarA L5 loop, 2nd view</scene>. The loop is shown in purple, and the residues of the loop as purple ball-and-sticks. The rest of the enzyme is visualized in blue space-fill representation. | ||
| + | |||
| + | L5 loop conformation types: | ||
| + | *<scene name='98/982642/Shorter-type_l5/1'>Shorter-type L5 loop</scene> of DbeA (green), DhaA (gray), DmxA (pink) and DmmA (yellow) is depicted. | ||
| + | *<scene name='98/982642/Longer-type_l5/1'>Longer-type L5 loop</scene> of HanR (gold), LinB (brown), DccA (medium-orchid) and DpaA (salmon). | ||
| + | *<scene name='98/982642/Dmmara_l5_loop/2'>DmmarA-type L5 loop</scene> (purple). | ||
| + | *<scene name='98/982642/All_l5_loops/2'>The superposition of all L5 loops</scene> is displayed. | ||
| + | |||
| + | Diverse dimerization modes in the HLD family: | ||
| + | *<scene name='98/982642/Superpositionall/1'>The superposition of all HLD dimers</scene>. | ||
| + | *<scene name='98/982642/Front-to-front/1'>Front-to-front interaction</scene> of DmxA (pink), DbeA (green) and DmmA (yellow). | ||
| + | *<scene name='98/982642/Cap-to-cap/1'>Cap-to-cap interaction</scene> of DpaA (salmon) and HanR (gold). | ||
| + | *<scene name='98/982642/Back-to-back/1'>Back-to-back dimerization interface</scene> of DmmarA (blue). | ||
| + | *<scene name='98/982642/Bottom-to-bottom/1'>Bottom-to-bottom interaction</scene> of DccA (medium-orchid). | ||
| + | |||
| + | <b>References</b><br> | ||
| + | <references/> | ||
| + | </StructureSection> | ||
| + | __NOEDITSECTION__ | ||
Current revision
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.