Cas9 Sandbox
From Proteopedia
(Difference between revisions)
| (One intermediate revision not shown.) | |||
| Line 1: | Line 1: | ||
== Function == | == Function == | ||
<StructureSection load='4un3' size='350' frame='true' side='right' caption='4un3 of Streptococcus pyogenes' scene='Insert optional scene name here' > | <StructureSection load='4un3' size='350' frame='true' side='right' caption='4un3 of Streptococcus pyogenes' scene='Insert optional scene name here' > | ||
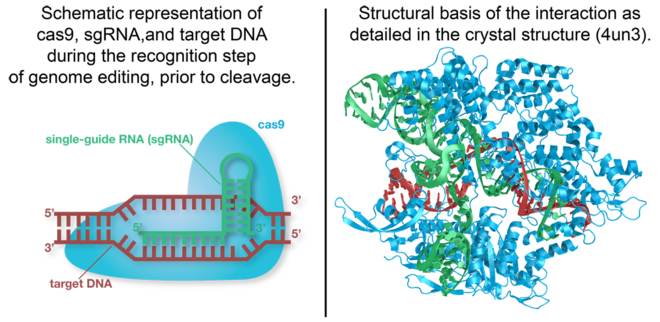
| - | [[Cas9]] is the RNA-guided [[DNA endonuclease]] used by the CRISPR (clustered regularly interspaced short palindromic repeats)-associated systems to generate double-strand DNA breaks in the invading DNA during an adaptive bacterial immune response. Three different types of CRISPR mechanisms have been discovered, however, only type II CRISPR systems are heavily researched. In vivo, [[Cas9]] requires CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) to guide the endonuclease toward invading DNA based on the complementary sequence recognition of these RNAs. Cas9 then targets and cleaves foreign DNA to interfere with viral replication and integration into the host's genome. The CRISPR-associated endonuclease <scene name='71/714945/ | + | [[Cas9]] is the RNA-guided [[DNA endonuclease]] used by the CRISPR (clustered regularly interspaced short palindromic repeats)-associated systems to generate double-strand DNA breaks in the invading DNA during an adaptive bacterial immune response. Three different types of CRISPR mechanisms have been discovered, however, only type II CRISPR systems are heavily researched. In vivo, [[Cas9]] requires CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) to guide the endonuclease toward invading DNA based on the complementary sequence recognition of these RNAs. Cas9 then targets and cleaves foreign DNA to interfere with viral replication and integration into the host's genome. The CRISPR-associated endonuclease <scene name='71/714945/Dna_rna_cas9_cartoon/1'>Cas9</scene> has been exploited for use in genome editing systems. In such systems, an engineered single-guide RNA (sgRNA) is used to perform the function of crRNA-tracRNA complex to target double-stranded breaks in genomic DNA. Depending on what repair pathway is triggered, often dictated by the inclusion of additional engineered components, the targeted site either is disrupted or incorporates additional genetic sequences. <ref>PMID:24906146</ref> |
| Line 8: | Line 8: | ||
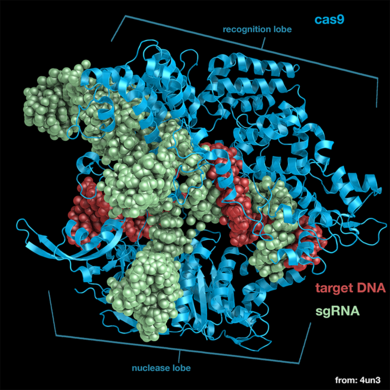
Cas9 is a structurally bilobed, containing specific domains for the recognition of target DNA and nucleases to cleave DNA strands. Each of these lobes contain three major units essential for a functional endonuclease, these are the essential <scene name='71/714945/Lobes_domain_of_cas9/5'>domains</scene>. | Cas9 is a structurally bilobed, containing specific domains for the recognition of target DNA and nucleases to cleave DNA strands. Each of these lobes contain three major units essential for a functional endonuclease, these are the essential <scene name='71/714945/Lobes_domain_of_cas9/5'>domains</scene>. | ||
| - | The recognition lobe (REC) contains a long bridge helix, the REC-1 domain and the REC-2 domain. It is also the least conserved lobe through the types of Cas9. The long <scene name='71/714945/Alpha_helix_bridge/1'>α-helix bridge</scene>, which is arginine-rich, is essential for recognizing single guide RNA-DNA complexes on target DNA. This structure has been shown to be conserved through Cas9 proteins. The <scene name='71/714945/Rec-1_domain/2'>REC-1</scene> domain contains 25 α-helixes and two β-sheets, and is crucial to the function of Cas9 by recognizing a specific motif termed the repeat:anti-repeat region of the single guide RNA and DNA complex. The <scene name='71/714945/Rec-2_domain/1'>REC-2</scene> domain contains six α-helix’s in a bundle but there is no current understanding of its function. | + | The <scene name='71/714945/Rec_domains/2'>recognition lobe</scene> (REC) contains a long bridge helix, the REC-1 domain and the REC-2 domain. It is also the least conserved lobe through the types of Cas9. The long <scene name='71/714945/Alpha_helix_bridge/1'>α-helix bridge</scene>, which is arginine-rich, is essential for recognizing single guide RNA-DNA complexes on target DNA. This structure has been shown to be conserved through Cas9 proteins. The <scene name='71/714945/Rec-1_domain/2'>REC-1</scene> domain contains 25 α-helixes and two β-sheets, and is crucial to the function of Cas9 by recognizing a specific motif termed the repeat:anti-repeat region of the single guide RNA and DNA complex. The <scene name='71/714945/Rec-2_domain/1'>REC-2</scene> domain contains six α-helix’s in a bundle but there is no current understanding of its function. |
The nuclease lobe (NUC) contains a RuvC domain, HNH domain and the PAM-interacting domain (PI). The RuvC domain is comprised of three RuvC motifs that are made up of two-stranded antiparallel β-sheets, and six-stranded β-sheets, which are flanked by nine α-helices. The <scene name='71/714945/Ruvc_domain/1'>RuvC nuclease</scene> cleaves the non-complementary, single stranded DNA. This nuclease is active through the use of four catalytic residues shown in the previous scene. The <scene name='71/714945/Hnh_domain/3'>HNH domain</scene> is composed of a two-stranded antiparallel β-sheet which is flanked by four α-helixes and cleaves the complementary strand of target DNA. The four catalytic residues of this nuclease domain are highlighted in red. The <scene name='71/714945/Pi/1'>PI</scene> is made up of seven α-helixes and numerous strand-varying antiparallel β-sheets which recognize the PAM sequence on the non-complementary target DNA strand.<ref>PMID:24529477</ref> | The nuclease lobe (NUC) contains a RuvC domain, HNH domain and the PAM-interacting domain (PI). The RuvC domain is comprised of three RuvC motifs that are made up of two-stranded antiparallel β-sheets, and six-stranded β-sheets, which are flanked by nine α-helices. The <scene name='71/714945/Ruvc_domain/1'>RuvC nuclease</scene> cleaves the non-complementary, single stranded DNA. This nuclease is active through the use of four catalytic residues shown in the previous scene. The <scene name='71/714945/Hnh_domain/3'>HNH domain</scene> is composed of a two-stranded antiparallel β-sheet which is flanked by four α-helixes and cleaves the complementary strand of target DNA. The four catalytic residues of this nuclease domain are highlighted in red. The <scene name='71/714945/Pi/1'>PI</scene> is made up of seven α-helixes and numerous strand-varying antiparallel β-sheets which recognize the PAM sequence on the non-complementary target DNA strand.<ref>PMID:24529477</ref> | ||
Current revision
Function
| |||||||||||