User:Camille Gaudet/Sandbox 1
From Proteopedia
< User:Camille Gaudet(Difference between revisions)
(New page: ==Your Heading Here (maybe something like 'Structure')== <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> This is a default text for you...) |
|||
| (131 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | = | + | ='''Glucose-dependent Insulinotropic Polypeptide Receptor'''= |
| - | <StructureSection load= | + | <StructureSection load=7RA3 'size='350' frame='true'side='right' caption='Glucose-dependent Insulinotropic Polypeptide, 7RA3' scene='10/1037488/Gip-r_noligand_noantibody/4'> |
| - | + | ||
| - | + | ||
| - | == Function == | ||
| - | == | + | == Introduction == |
| + | === History === | ||
| + | Investigation of pancreatic hormones began at the turn of the 20th century. In 1902, Bayliss and Starling discover a pancreatic [https://en.wikipedia.org/wiki/Secretin secretin] involved in regulation of water homeostasis, giving rise to interest in pancreatic hormones. Shortly after, in 1906, Moore et al. hypothesize involvement of [https://en.wikipedia.org/wiki/Gastrointestinal_hormone gastrointestinal hormone extracts] in maintenance of the endocrine pancreas. Jean La Barre purifies glucose-lowering gut extracts in 1929 and characterizes them as [https://en.wikipedia.org/wiki/Incretin incretins], which is short for intestine secretion insulin. Finally, in 1984, [https://en.wikipedia.org/wiki/Glucose-dependent_insulinotropic_polypeptide gastric inhibitory peptide] (GIP) is isolated from porcupine intestine. Although GIP was initially characterized for its gastric inhibitory effects (hence the name), it was also shown that the polypeptide played an integral role in [https://en.wikipedia.org/wiki/Insulin_signal_transduction_pathway insulin signaling and secretion]. Interestingly, it was found that the effect of GIP on insulin levels was still seen in its absence, hinting toward the presence of an additional incretin, which has now been classified as [https://en.wikipedia.org/wiki/Glucagon-like_peptide-1 glucagon-like peptide-1] (GLP-1).<ref name ='Seino'>DOI:10.1111/j.2040-1124.2010.00022.x</ref> | ||
| - | == | + | === Biological Role === |
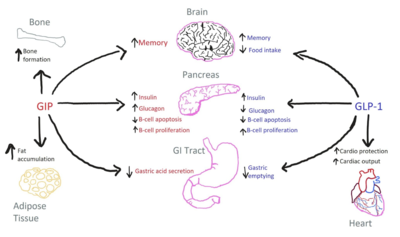
| + | [[Image:GIP-GLP1-Use.png|400 px|right|thumb|Figure 1. The biological roles of GIP and and GLP-1, incretin hormones.]] The GIP receptor (GIPR) helps facilitate the transport of glucose into/out of the cell through the stimulation of insulin secretion. <ref name='Sun'>PMID:35333651</ref>. GIPR is a type of [https://en.wikipedia.org/wiki/G_protein-coupled_receptor G-Protein Coupled Receptor] (GPCR), and its natural ligand, GIP, serves as an initiator of a cellular signaling cascade, thereby activating adenylyl cyclase and increasing cAMP levels. Subsequently, insulin secretion is stimulated. [https://en.wikipedia.org/wiki/Insulin Insulin], a peptide hormone, is secreted by the pancreas in response to glucose ingestion, allowing intake of glucose into the cell via the [https://en.wikipedia.org/wiki/GLUT2 Glut2] transporter. Notably, GIP, as well as GLP-1, serve a multitude of biological roles (Figure 1) other than insulin signaling.<ref name ='Seino'>DOI:10.1111/j.2040-1124.2010.00022.x</ref> | ||
| - | == Structural highlights == | ||
| - | + | == General Structure == | |
| + | === The Receptor and G Protein (GIPR) === | ||
| + | <scene name='10/1037488/Gip-r_noligand_noantibody/9'>GIP-R without GIP</scene> appears as a typical G-Protein Coupled Receptor, containing a <scene name='10/1037488/Gip-r_transmembrane/1'>transmembrane domain</scene> with seven helical passes and an <scene name='10/1037488/Gip-r_extracellular/1'>extracellular domain</scene>, where the ligand binds. These two domains comprise the <scene name='10/1037488/Gip-r_receptor-centered/6'>receptor</scene>. Intracellularly, the receptor is bound to a G-Protein which is comprised by the <scene name='10/1037488/Gip-r_galpha-centered/4'>G Alpha (Gs⍺iN18)</scene>, <scene name='10/1037488/Gip-r_gbeta-centered/5'>G Beta (Gβ)</scene>, and <scene name='10/1037488/Gip-r_ggamma-centered/3'>G Gamma (Gγ)</scene> subunits, where the Beta and Gamma subunits typically [https://en.wikipedia.org/wiki/Dimerization_(chemistry) dimerize]. Upon binding of GIP to the receptor, the intracellular signaling cascade is initiated through Gs⍺iN18 stimulation of adenylyl cyclase and increased [cAMP]. | ||
| + | |||
| + | === Glucose-dependent Insulinotropic Polypeptide === | ||
| + | The <scene name='10/1037488/Gip-bound/4'>GIP ligand</scene> binds to its receptor extracellularly, inserting itself [https://en.wikipedia.org/wiki/N-terminus N-terminus] down into the transmembrane and extracellular domains of the GIP receptor. The polypeptide is 42 residues in length and takes on a helical structure. | ||
| + | |||
| + | === Active Site === | ||
| + | A crucial binding interaction within the active site occurs between <scene name='10/1037488/Gip-y1-intrxn-still/8'>Tyr1 of GIP</scene> and receptor residues W296 and Q224. The hydroxyl of the tyrosine residue forms a [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond] to the amide of the glutamine, and the aromaticity of both the tyrosine and tryptophan residues results in [https://en.wikipedia.org/wiki/Stacking_(chemistry) pi stacking] between them. In GIP [https://en.wikipedia.org/wiki/Agonist agonist] diabetes medications, conservation of the Tyr1 residue can determine the drug’s efficacy in initiating a GIP-like response.<ref name="Sun">PMID: 35333651</ref> | ||
| + | |||
| + | |||
| + | == Associated Diseases == | ||
| + | In individuals with [https://en.wikipedia.org/wiki/Diabetes diabetes], insulin is improperly under-secreted or unresponsive to elevated blood glucose levels. Considering its role as the initial metabolite in [https://en.wikipedia.org/wiki/Glycolysis glycolysis], when glucose is not effectively transported into the cell, it renders the body unable to successfully execute energy production through [https://en.wikipedia.org/wiki/Cellular_respiration cellular respiration] (Glycolysis --> the Krebs Cycle --> the Electron Transport Chain --> ATP synthesis). | ||
| + | |||
| + | == Medical Relevance == | ||
| + | Many anti-diabetes drugs exist for patients with both Type I and Type II diabetes, including a new dual GIP and GLP-1 agonist known as [https://en.wikipedia.org/wiki/Tirzepatide Tirzepatide] (Brand Name: Mounjaro, Manufacturer: [https://en.wikipedia.org/wiki/Eli_Lilly_and_Company Lilly]). Tirzepatide is capable of inducing the effects of both GIP and GLP-1 cellular signalling by mimicking each ligand and binding to their respective receptor. Increased presence (or, the apparent increased presence) of GIP and GLP-1 leads to enhanced stimulation of insulin secretion, thereby reducing blood glucose levels.<ref name="Chavda">PMID: 35807558</ref> | ||
| + | |||
| + | === Tirzepatide Structure=== | ||
| + | Structurally, as a dual agonist, <scene name='10/1037488/Tirz-bound/3'>Tirzepatide</scene> closely resembles GIP and GLP-1. The sequence alignment of all three polypeptides showcases the highly integrated nature of both GIP and GLP-1 into the amino acid sequence of Tirzepatide with very few alterations (Figure 2). [[Image:GIPGLPTirz.png|700 px|right|thumb|Figure 2. Sequence alignment of GLP-1, GIP, and Tirzepatide. Residues shown in black are found in all three amino acid sequences. Pink or red coloration denotes residues that are unique to GIP or GLP-1, respectively. The sequence of tirzepatide is colored accordingly, and residues differing from both GIP and GLP-1 are highlighted in blue. Residues that differ across all three structures are boxed.]] The <scene name='10/1037488/Tirz-y1conserved/2'>conserved Tyr1 residue</scene> allows for simulation of a highly similar interaction between Tirzepatide and the GIP receptor. Had this been altered, Tirzepatide would not nearly bind with as high of an affinity for GIPR. The Tyr1 residue facilitates strong contacts with the TMD core. Several <scene name='10/1037488/Tirz-bound-differences/3'>unique residues in Tirzepatide</scene> do not align with either the sequence of GIP or GLP-1 and are located toward the C-terminus end of the structure. They include Ala21, Gln24, Ile27, and Gly30. The <scene name='10/1037488/Gip-differences/6'>corresponding GIP residues</scene> are Asp21, Asn24, Leu27, and Lys30. Interestingly, these residues do not interact with the receptor, thus their mutation doesn't alter the overall binding affinity of Tirzepatide to GIPR.<ref name="Sun">PMID: 35333651</ref> Other significant structural modifications were made to maximize GIPR-Tirz interactions.The AIB (alpha-aminoisobutyric) residues (Figure 2) prevent degradation of the polypeptide by peptidases such as DPP-4.<ref name="Zhao">DOI:10.1038/s41467-022-28683-0</ref> | ||
| + | Because Tirzepatide was modeled after two distinct polypeptides, its binding to each of the receptors has hallmark differences. The Tirz-GIPR complex is rotated ~8.3º compared to Tirz-GLP-1R, with the C-terminus translated closer to the TMD core. ECL1 interactions with Tirzepatide are diminished in GIPR due to the presence of several <scene name='10/1037488/Gip-r_receptor-centered/7'>repeating proline residues (P195, P197, and P199)</scene>. Fortunately, the ɑ-helical extension provides another recognition point for Tirzepatide; a hydrogen bond between <scene name='10/1037488/Tirz-y10/1'>Y10 (Tirz) and Q138 (GIPR)</scene> and stacking between K16 (Tirz) and F127 (GIPR). Primary interactions between the receptor and Tirzepatide occur within the first 27 residues (Figure 3).<ref name="Zhao">DOI:10.1038/s41467-022-28683-0</ref> | ||
| + | [[Image:GIPRTirz-binding.png|700 px|right|thumb|Figure 3. Amino acid sequence of Tirzepatide and the type of interaction it forms with the GIP receptor. Salt bridges (blue), hydrogen bonds (red), pi stacking (orange), and Van der Waals (gray) interactions are highlighted.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Future Directions=== | ||
| + | In addition to treating diabetes, GIP and GLP-1 agonists have grown in recently popularity due to their weight loss inducing effects. The dual agonism of Tirzepatide is thought to be more effective than agonists of GLP-1/GIP alone, though no published studies have formally confirmed this hypothesis. Further research into other pancreatic hormones is required to determine additional routes of insulin stimulation. | ||
| + | |||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | <ref name="Chavda">PMID: 35807558</ref> | ||
| + | <ref name ='Seino'>DOI:10.1111/j.2040-1124.2010.00022.x</ref> | ||
| + | <ref name="Sun">PMID: 35333651</ref> | ||
| + | <ref name="Zhao">DOI:10.1038/s41467-022-28683-0</ref> | ||
<references/> | <references/> | ||
| + | |||
| + | ==Student Contributors== | ||
| + | *Camille Gaudet | ||
| + | *Sara Kalkhoff | ||
Current revision
Glucose-dependent Insulinotropic Polypeptide Receptor
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Seino Y, Fukushima M, Yabe D. GIP and GLP-1, the two incretin hormones: Similarities and differences. J Diabetes Investig. 2010 Apr 22;1(1-2):8-23. PMID:24843404 doi:10.1111/j.2040-1124.2010.00022.x
- ↑ 2.0 2.1 2.2 2.3 Sun B, Willard FS, Feng D, Alsina-Fernandez J, Chen Q, Vieth M, Ho JD, Showalter AD, Stutsman C, Ding L, Suter TM, Dunbar JD, Carpenter JW, Mohammed FA, Aihara E, Brown RA, Bueno AB, Emmerson PJ, Moyers JS, Kobilka TS, Coghlan MP, Kobilka BK, Sloop KW. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2116506119. PMID:35333651 doi:10.1073/pnas.2116506119
- ↑ 3.0 3.1 Chavda VP, Ajabiya J, Teli D, Bojarska J, Apostolopoulos V. Tirzepatide, a New Era of Dual-Targeted Treatment for Diabetes and Obesity: A Mini-Review. Molecules. 2022 Jul 5;27(13):4315. PMID:35807558 doi:10.3390/molecules27134315
- ↑ 4.0 4.1 4.2 Zhao F, Zhou Q, Cong Z, Hang K, Zou X, Zhang C, Chen Y, Dai A, Liang A, Ming Q, Wang M, Chen LN, Xu P, Chang R, Feng W, Xia T, Zhang Y, Wu B, Yang D, Zhao L, Xu HE, Wang MW. Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors. Nat Commun. 2022 Feb 25;13(1):1057. PMID:35217653 doi:10.1038/s41467-022-28683-0
Student Contributors
- Camille Gaudet
- Sara Kalkhoff