We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1f8a
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
<jmolCheckbox> | <jmolCheckbox> | ||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/f8/1f8a_consurf.spt"</scriptWhenChecked> | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/f8/1f8a_consurf.spt"</scriptWhenChecked> | ||
| - | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/ | + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> |
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1f8a ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1f8a ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
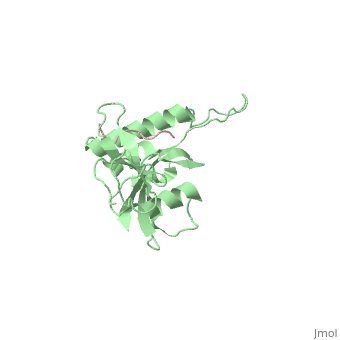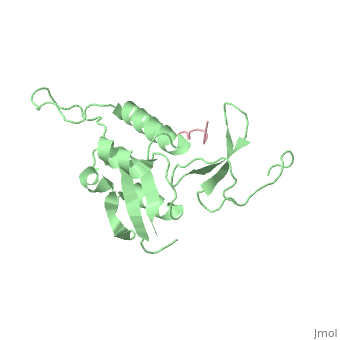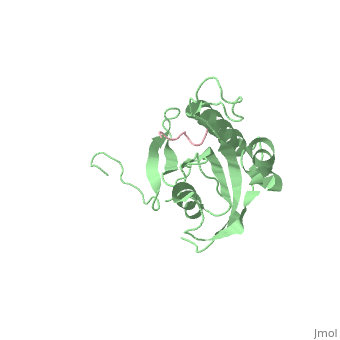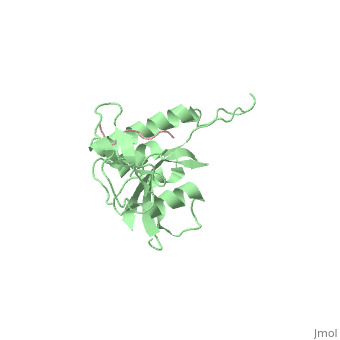
| + | Pin1 contains an N-terminal WW domain and a C-terminal peptidyl-prolyl cis-trans isomerase (PPIase) domain connected by a flexible linker. To address the energetic and structural basis for WW domain recognition of phosphoserine (P.Ser)/phosphothreonine (P. Thr)- proline containing proteins, we report the energetic and structural analysis of a Pin1-phosphopeptide complex. The X-ray crystal structure of Pin1 bound to a doubly phosphorylated peptide (Tyr-P.Ser-Pro-Thr-P.Ser-Pro-Ser) representing a heptad repeat of the RNA polymerase II large subunit's C-terminal domain (CTD), reveals the residues involved in the recognition of a single P.Ser side chain, the rings of two prolines, and the backbone of the CTD peptide. The side chains of neighboring Arg and Ser residues along with a backbone amide contribute to recognition of P.Ser. The lack of widespread conservation of the Arg and Ser residues responsible for P.Ser recognition in the WW domain family suggests that only a subset of WW domains can bind P.Ser-Pro in a similar fashion to that of Pin1. | ||
| + | |||
| + | Structural basis for phosphoserine-proline recognition by group IV WW domains.,Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP Nat Struct Biol. 2000 Aug;7(8):639-43. PMID:10932246<ref>PMID:10932246</ref> | ||
| + | |||
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
| + | <div class="pdbe-citations 1f8a" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
Current revision
STRUCTURAL BASIS FOR THE PHOSPHOSERINE-PROLINE RECOGNITION BY GROUP IV WW DOMAINS
| |||||||||||
Categories: Homo sapiens | Large Structures | Bowman ME | Hunter T | Lu KP | Noel JP | Verdecia MA