We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Mandy Bechman/Sandbox 1
From Proteopedia
< User:Mandy Bechman(Difference between revisions)
| (48 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
=H. sapiens Glucose-dependent Insulinotropic Polypeptide= | =H. sapiens Glucose-dependent Insulinotropic Polypeptide= | ||
| + | <StructureSection load='7ra3' size='400' side='right' caption='GIP Structure' scene='10/1038867/Structure_overview/10'> | ||
==Introduction== | ==Introduction== | ||
| + | Glucose-dependent insulinotropic polypeptide is a gastric inhibitory polypeptide, composed of 42 amino acids. It is categorized as an incretin, which is a gut-based hormone that is secreted upon the ingestion of food. It is found in K cells in the upper small intestines of humans.<ref name="Mayendraraj"/> | ||
===History=== | ===History=== | ||
| + | The idea of incretin, and subsequently GIP, began in 1902 after the discovery of secretin by Bayliss and Starling. Inspired by their discovery, Moore et al. hypothesized that there must be some kind of gut hormone that regulates the endocrine pancreas. His experiments showed that gut extracts in patients with diabetes have reduced amounts of sugars in their urine as a result of endocrine stimulation. In 1929, a French scientist named La Barre was able to purify incretin (INtestine seCREtion INsulin) from gut extracts. The idea of incretin slowly lost attention until the 1960s when radioimmunoassay became available to measure incretin levels. GIP and GLP-1 (glucagon-like peptide-1) were the two gut based hormones that were proven to act as incretins. GIP was found to be located in the K cells of the upper small intestines in humans. At first, it was titled a gastric inhibitory polypeptide because of its ability to inhibit gastric acid secretion after being isolating from porcine intestines. Later, they discovered that when GIP was administered in healthy volunteers, it stimulated insulin secretion by acting directly on pancreatic islets. As a result of these discoveries, they renamed it glucose-dependent insulinotropic polypeptide, becoming the first known incretin.<ref name="Seino"/> | ||
| + | |||
| + | |||
==Structure Overview== | ==Structure Overview== | ||
| - | <scene name='Structure_overview> | + | ===G-proteins=== |
| - | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4020673/ | + | <scene name='10/1038867/Structure_overview/10'>GIP</scene> is a G-coupled receptor [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4020673/ GPCR]. It is a 7 helical transmembrane domain with the ligand in dark blue and the helices in light blue. It contains alpha, beta, and gamma <scene name='10/1038867/Structure_overview/11'>subunits</scene> located in the intracellular domain. The G alpha subunit makes interactions with the beta and gamma subunits that when bound to ATP, activates the G alpha protein and is released by the other subunits to bind to target, i.e. adenylyl cycle in order to activate the process of insulin secretion.<ref name="Seino"/> |
| - | < | + | |
| - | + | ||
| - | + | ||
== Function == | == Function == | ||
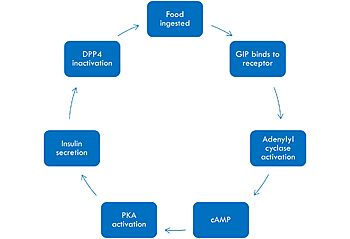
| - | < | + | [[Image:GIPmech.jpg|350 px|left|thumb|Mechanism of GIP activation and deactivation.]] |
| + | GIP is a vital hormone for the daily function of humans. It is involved in increased bone formation as well as increased lipogenesis, which is the conversion and storage of fats as triglycerides in adipocyte tissues. In the brain, GIP acts increase brain cell proliferation, which is very important for early stage development in children. In the pancreas, GIP acts to increase insulin production, increase glucagon levels, increase beta cell proliferation as well as decrease beta cell apoptosis levels. In the GI tract, this hormone acts to decrease gastric acid secretion.<ref name="Seino"/> | ||
| + | ===Mechanism=== | ||
| + | The mechanism of GIP begins with the ingestion of food. This stimulates GIP to bind to its receptor. Once bound, the activated G alpha protein moves laterally in the membrane in order to activate its target, adenylyl cyclase. Once adenylyl cyclase is activated, it catalyzes the formation of cyclic AMP (cAMP). This molecule activates Protein Kinase A (PKA), which signals the secretin of insulin. After a few minutes of active signaling, this hormone is recognized and inactivated by a peptidase called DPP-4 (dipeptidyl peptidase-4) by cleaving the first two amino acids.<ref name="Mayendraraj"/> | ||
| + | ===Diseases=== | ||
| + | Malfunction of the GIP protein can result in some serious life-threatening diseases. Overexpression of GIP can result in obesity by promoting fat disposition in adipocyte tissues as well as an inability to regulate food intake and control appetite. Underexpression of GIP can result in diabetes, a disease associated with the lack of insulin biosynthesis, which is vital for energy production. | ||
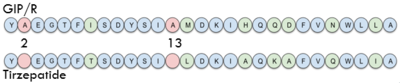
| - | == | + | ====Tirzepatide==== |
| + | One of the most promising treatments to GIP malfunction is <scene name='10/1038867/Tirzepatide/2'>Tirzepatide</scene>, also known as Mounjaro, created by Eli Lily. It is an antidiabetic as well as weight loss drug. The structure of Tirzepatide functions as an incretin receptor agonist to GIP by activating GIP's receptor and producing insulin. | ||
| + | [[Image:Sequence.png|400 px|left|thumb|Sequence comparison of the first 28 residues of GIP and Tirzepatide.]] | ||
| - | == Relevance == | ||
== Structural highlights == | == Structural highlights == | ||
| + | === Binding/Active Site of GIPR with GIP === | ||
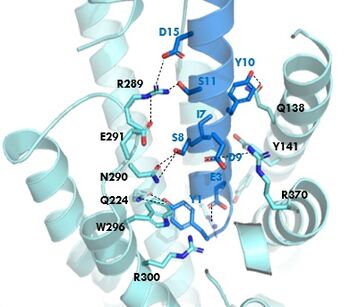
| + | [[Image:GIP_hydrogen_bonds.jpg|350 px|right|thumb|GIPR (light blue) and GIP (dark blue) residue interactions]] | ||
| + | The <scene name='10/1038815/Overview/7'>binding site</scene> of GIP with the GIP receptor (GIPR) is where the N-term of GIP binds with the transmembrane domain of the GIPR. The first interaction formed with GIPR and the N-term of GIP is a hydrogen bond between Tyrosine 1 (Y1) and Glutamine 224 (Q224) to activate the G-protein to start sending signals to the cell. | ||
| + | Many other <scene name='10/1038815/Active_site/3'>residues</scene> within the binding site are forming hydrogen bonds and hydrophobic interactions between the ligand and the receptor. The N-term binds more strongly than the C-term and there are many different residues contributing to this, including, Tyrosine 1 (Y1) and Tryptophan (W296) are forming aromatic interactions. Serine 8 (S8) and Asparagine 290 (N290) are forming two hydrogen bonds with each other. Aspartate 9 (D9) is forming another hydrogen bond with Arginine 370 (R370)<ref name="Sun"/>. All of these interactions are responsible for the binding affinity, the strength of the attraction, between the two proteins. These hydrogen bonds lead to the activation of cell signaling and when this binding is somehow disrupted, that is what causes different diseases like diabetes. | ||
| + | === Binding/Active Site of GIPR with Tirzepatide === | ||
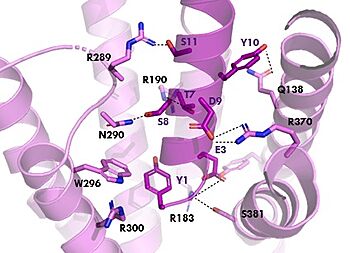
| + | [[Image:TZ_hydrogen_bonds.jpg|350 px|right|thumb|GIPR (pink) and Tirzepatide (dark pink) residue interactions]] | ||
| + | The <scene name='10/1038815/Tirzepatide_active_site/4'>binding site</scene> with Tirzepatide is the same as GIP with the N-term binding to the transmembrane domain and activating cellular signaling.The <scene name='10/1038815/Tirzepatide_residues/7'>residues</scene> are fairly similar just in a different conformations, which is allowing for more hydrogen bonding. In Tirzepatide, the most noticeable conformational change is the Tyrosine 1 (Y1) residue. It is now facing up towards Arginine 190 (R190) where they can now form a new hydrogen bond that was not present in GIP. The more hydrogen bonding there is between molecules, the stronger the binding between the two molecules will be. This can lead to the conclusion that GIPR has a higher binding affinity for Tirzepatide than GIP itself<ref name="Sun"/>. | ||
| + | === Isoleucine vs. Threonine === | ||
| + | In GIP there is an Isoleucine present <scene name='10/1038815/Gip_ile7/3'>(I7)</scene>. Isoleucine is a branched residue that is very hydrophobic. This residue is also present in the N-term where the important interactions are occuring. In Tirzepatide, this seventh residue has been changed to a Threonine residue <scene name='10/1038815/Tirzepatide_thr7/3'>(T7)</scene>. Threonine is also a branched residue, but it contains a hydroxyl group making it hydrophilic, allowing it to make hydrogen bonds. Threonine 7 (T7) is also making a hydrogen bone with the Arginine 190 (R190) residue. This extra hydrogen bond in Tirzepatide is also responsible for a higher binding affinity for the drug than GIP itself. | ||
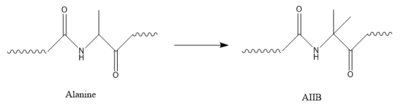
| + | ===AIB Mutation=== | ||
| + | [[Image:AlatoAIB.png|400 px|left|thumb|Structure of alanine converted to AIB.]] | ||
| + | In GIP, residue 2 is an <scene name='10/1038867/Alanine_2_-_gip/3'>alanine</scene> but in Tirzepatide, the residue changes to <scene name='10/1038867/Tirzepatide_-_aib/2'>AIB</scene>. Residue 2 is where DPP4 recognizes GIP in order to cleave and inactivate the hormone. When the residue is changed, it is no longer recognized by DPP4 because AIB is a noncoding residue, resulting in continuous signaling to the cell. | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | <ref name=" | + | <ref name="Seino">PMID:24843404</ref>. |
| - | <ref name="Mayendraraj>PMID:35065096</ref>. | + | <ref name="Mayendraraj">PMID:35065096</ref>. |
| + | <ref name="Sun">PMID:35333651</ref> | ||
<references/> | <references/> | ||
| + | |||
| + | ==Student Contributors== | ||
| + | Mandy M. Bechman | ||
| + | Chloe A. Tucker | ||
Current revision
H. sapiens Glucose-dependent Insulinotropic Polypeptide
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Mayendraraj A, Rosenkilde MM, Gasbjerg LS. GLP-1 and GIP receptor signaling in beta cells interactions and co-stimulation. Peptides. 2022 May;151:170749. PMID:35065096 doi:10.1016/j.peptides.2022.170749
- ↑ 2.0 2.1 2.2 2.3 Seino Y, Fukushima M, Yabe D. GIP and GLP-1, the two incretin hormones: Similarities and differences. J Diabetes Investig. 2010 Apr 22;1(1-2):8-23. PMID:24843404 doi:10.1111/j.2040-1124.2010.00022.x
- ↑ 3.0 3.1 3.2 Sun B, Willard FS, Feng D, Alsina-Fernandez J, Chen Q, Vieth M, Ho JD, Showalter AD, Stutsman C, Ding L, Suter TM, Dunbar JD, Carpenter JW, Mohammed FA, Aihara E, Brown RA, Bueno AB, Emmerson PJ, Moyers JS, Kobilka TS, Coghlan MP, Kobilka BK, Sloop KW. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2116506119. PMID:35333651 doi:10.1073/pnas.2116506119
Student Contributors
Mandy M. Bechman Chloe A. Tucker