This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Brynn Baker/Sandbox1
From Proteopedia
< User:Brynn Baker(Difference between revisions)
m |
|||
| (8 intermediate revisions not shown.) | |||
| Line 6: | Line 6: | ||
== Amylin == | == Amylin == | ||
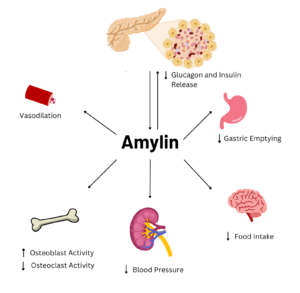
| - | [[Image:AmylinFlowchart.png|300 px|right|thumb|Figure 1. Effects of Amylin in Humans]] | + | [[Image:AmylinFlowchart.png|300 px|right|thumb|Figure 1. Effects of Amylin in Humans. Image generated using ''Biorender''.]] |
Amylin is a neuroendocrine hormone that is synthesized with insulin in the [https://en.wikipedia.org/wiki/Beta_cell beta cells] of pancreatic islets. It can cross the [https://en.wikipedia.org/wiki/Blood%E2%80%93brain_barrier blood-brain barrier] and regulates glucose homeostasis via inhibiting gastric emptying, inhibiting the release of glucagon, and inducing meal-ending satiety<ref name="Hay">PMID:26071095</ref>. In doing so, it prevents spikes in blood glucose and overeating, making it a suitable target for [https://en.wikipedia.org/wiki/Type_2_diabetes Type 2 Diabetes] treatments and therapies. Since Type 2 Diabetes is a major risk factor for [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's Disease], as Type 2 Diabetes cases continue to increase, there will likely be a spike in Alzheimer’s Disease as well<ref name="Grizzanti">PMID:30282360</ref>. Therefore, it is vital that amylin, its receptor, and analogs, such as pramlintide, are understood to aid in rational drug design. | Amylin is a neuroendocrine hormone that is synthesized with insulin in the [https://en.wikipedia.org/wiki/Beta_cell beta cells] of pancreatic islets. It can cross the [https://en.wikipedia.org/wiki/Blood%E2%80%93brain_barrier blood-brain barrier] and regulates glucose homeostasis via inhibiting gastric emptying, inhibiting the release of glucagon, and inducing meal-ending satiety<ref name="Hay">PMID:26071095</ref>. In doing so, it prevents spikes in blood glucose and overeating, making it a suitable target for [https://en.wikipedia.org/wiki/Type_2_diabetes Type 2 Diabetes] treatments and therapies. Since Type 2 Diabetes is a major risk factor for [https://en.wikipedia.org/wiki/Alzheimer%27s_disease Alzheimer's Disease], as Type 2 Diabetes cases continue to increase, there will likely be a spike in Alzheimer’s Disease as well<ref name="Grizzanti">PMID:30282360</ref>. Therefore, it is vital that amylin, its receptor, and analogs, such as pramlintide, are understood to aid in rational drug design. | ||
| Line 12: | Line 12: | ||
[[Image:Amylin_alignment_sequence.png|400 px|left|thumb|Figure 2. Amylin sequence alignment for human amylin, rat amylin, and pramlintide. Green represents fully conserved residues, pink represents positive to positive, red is nonpolar to nonpolar, and black is polar to nonpolar.]] | [[Image:Amylin_alignment_sequence.png|400 px|left|thumb|Figure 2. Amylin sequence alignment for human amylin, rat amylin, and pramlintide. Green represents fully conserved residues, pink represents positive to positive, red is nonpolar to nonpolar, and black is polar to nonpolar.]] | ||
| - | + | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
== Amylin Receptor == | == Amylin Receptor == | ||
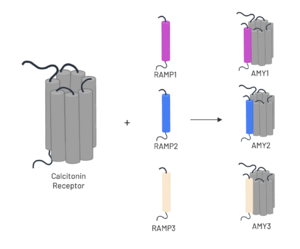
| - | The amylin receptor (AMYR) is the result of the heterodimerization of the <scene name='10/1037516/Ct/2'>calcitonin receptor</scene> and a RAMP, such as <scene name='10/ | + | The amylin receptor (AMYR) is the result of the heterodimerization of the <scene name='10/1037516/Ct/2'>calcitonin receptor</scene> and a RAMP, such as <scene name='10/1037520/Ramp3/5'>RAMP3</scene>. The patterns of peptide interaction between CT and AMYR are very similar overall, but amylin has a higher affinity for AMYR1 and AMYR3 than AMYR2<ref name="Cao">PMID:35324283</ref>. |
| - | [[Image:AMYR.png|300 px|left|thumb|Figure 3. Heterodimerization of CT and RAMPs]] | + | [[Image:AMYR.png|300 px|left|thumb|Figure 3. Heterodimerization of CT and RAMPs. Image generated using ''Biorender''.]] |
== Calcitonin Receptor and G-alpha Interactions == | == Calcitonin Receptor and G-alpha Interactions == | ||
| - | The calcitonin receptor forms several highly conserved interactions with the Gɑ subunit. The <scene name='10/1037520/Calc_and_galpha_nande/ | + | The calcitonin receptor forms several highly conserved interactions with the Gɑ subunit. The <scene name='10/1037520/Calc_and_galpha_nande/3'>main chain of N396 on CT hydrogen bonds with the sidechain of E392 on Gɑ</scene>, as do the <scene name='10/1037520/Calc_and_galpha_qandi/3'>main chain of I248 and sidechain of Q384</scene>, respectively<ref name="Cao">PMID:35324283</ref>. |
== RAMPs == | == RAMPs == | ||
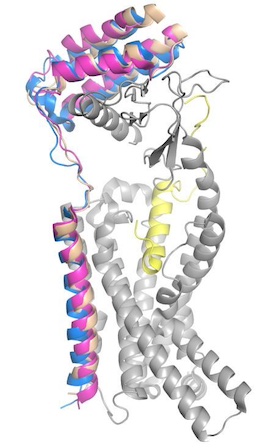
| - | To form the full AMYR complex, the calcitonin receptor must bind to one of three RAMPs: <scene name='10/1037520/Ramp1/1'>RAMP1</scene>, <scene name='10/1037520/Ramp2/2'>RAMP2</scene>, <scene name='10/1037520/Ramp3/ | + | To form the full AMYR complex, the calcitonin receptor must bind to one of three RAMPs: <scene name='10/1037520/Ramp1/1'>RAMP1</scene>, <scene name='10/1037520/Ramp2/2'>RAMP2</scene>, <scene name='10/1037520/Ramp3/5'>RAMP3</scene>. The three RAMPs have 31% sequence identity and bind very similarly to the CT, except RAMP2 has slightly looser packing<ref name="Cao">PMID:35324283</ref>. All three RAMPs serve the purpose of drastically increasing the CT’s binding affinity for amylin. RAMP3 is the version most heavily implicated in pramlintide targeting and Alzheimer’s Disease. |
[[Image:Rampstestmm2.jpg|300 px|right|thumb|Figure 4. Superimposed RAMP1 (pink), RAMP2 (blue), and RAMP3 (tan) in the human amylin receptor (CT is gray).]] | [[Image:Rampstestmm2.jpg|300 px|right|thumb|Figure 4. Superimposed RAMP1 (pink), RAMP2 (blue), and RAMP3 (tan) in the human amylin receptor (CT is gray).]] | ||
| Line 31: | Line 43: | ||
<scene name='10/1037520/Hydrophobic_pocket/6'>Y37</scene> on the C-terminus of the amylin peptide has extensive van der Waals interactions with the CT and RAMP3. The pi stacking and hydrophobic interactions increase the affinity and interactions between the amylin peptide, CT, and RAMP3, aiding in their association. | <scene name='10/1037520/Hydrophobic_pocket/6'>Y37</scene> on the C-terminus of the amylin peptide has extensive van der Waals interactions with the CT and RAMP3. The pi stacking and hydrophobic interactions increase the affinity and interactions between the amylin peptide, CT, and RAMP3, aiding in their association. | ||
=== Amidated C-Terminus === | === Amidated C-Terminus === | ||
| - | Y37 of amylin is amidated. The <scene name='10/ | + | Y37 of amylin is amidated. The <scene name='10/1037520/Amidated_cterm/2'>amide group</scene> forms a hydrogen bond with the backbone of S129 on the calcitonin receptor. All biological activity was lost when this amide group was experimentally removed<ref name="Cao">PMID:35324283</ref>. |
=== T6 === | === T6 === | ||
| Line 38: | Line 50: | ||
<scene name='10/1038871/R11/2'>R11</scene> is situated toward the N-terminal end of amylin’s main alpha helix and is packed against the residues at the N-terminus. This residue forms multiple interactions with various residues to keep the N-terminal end of the helix and the disulfide loop in place. R11 pulls on the backbones of C2 and C4 in the disulfide loop and interacts with the backbones of N3 and A8. It also attracts the hydroxyl group of Y372 of the calcitonin receptor. Furthermore, R11 stabilizes various nearby waters to form bridges between amylin and the receptor<ref name="Cao">PMID:35324283</ref>. | <scene name='10/1038871/R11/2'>R11</scene> is situated toward the N-terminal end of amylin’s main alpha helix and is packed against the residues at the N-terminus. This residue forms multiple interactions with various residues to keep the N-terminal end of the helix and the disulfide loop in place. R11 pulls on the backbones of C2 and C4 in the disulfide loop and interacts with the backbones of N3 and A8. It also attracts the hydroxyl group of Y372 of the calcitonin receptor. Furthermore, R11 stabilizes various nearby waters to form bridges between amylin and the receptor<ref name="Cao">PMID:35324283</ref>. | ||
=== R18 === | === R18 === | ||
| - | <scene name='10/ | + | <scene name='10/1037520/R18/1'>R18</scene> lies at the C-terminal end of the main alpha helix and can adopt two different configurations. The “upper” configuration provides stronger interactions with various residues on the calcitonin receptor and is potentially what causes the helix to terminate around R18. Specifically, R18 interacts with the sidechain of D97 and the backbones of F99, P100, and F102. The latter three residues may also form van der Waals interactions and pi stacking with each other<ref name="Cao">PMID:35324283</ref>. |
=== T9 Water Network === | === T9 Water Network === | ||
Threonine 9 is an essential residue for stabilization of amylin in the receptor, as its side chain is polar which allows it to hydrogen bond with other nearby polar groups, leading to extensive networks of interactions. T9 interacts with the <scene name='10/1038871/T9_main_chain/8'>main chains of Y191, M230, I380, and H381</scene> of the calcitonin receptor and many surrounding water molecules, but it also interacts with the <scene name='10/1038871/T9_network/4'>side chain atoms of S159, N194, S195, H226, N233, and Q383</scene>. All of these interactions create a very strong interaction between amylin and the receptor. The water network also helps stabilize the active receptor conformation. | Threonine 9 is an essential residue for stabilization of amylin in the receptor, as its side chain is polar which allows it to hydrogen bond with other nearby polar groups, leading to extensive networks of interactions. T9 interacts with the <scene name='10/1038871/T9_main_chain/8'>main chains of Y191, M230, I380, and H381</scene> of the calcitonin receptor and many surrounding water molecules, but it also interacts with the <scene name='10/1038871/T9_network/4'>side chain atoms of S159, N194, S195, H226, N233, and Q383</scene>. All of these interactions create a very strong interaction between amylin and the receptor. The water network also helps stabilize the active receptor conformation. | ||
| Line 47: | Line 59: | ||
The first human amylin analogue, <scene name='10/1037520/Pramlintide_overall/2'>pramlintide</scene>, was developed in 1995, and marked a significant advancement in the treatment of Type 2 Diabetes<ref name="Bower">PMID:27061187</ref>. As of 2024, it is the only FDA-approved drug for the treatment of Type 2 Diabetes using the AMYR as a target. Recent studies in rodent Alzheimer’s Disease models suggest that pramlintide reduces amyloid-beta plaques, making it a potential therapeutic target for Alzheimer’s Disease<ref name="Grizzanti">PMID:30282360</ref>. | The first human amylin analogue, <scene name='10/1037520/Pramlintide_overall/2'>pramlintide</scene>, was developed in 1995, and marked a significant advancement in the treatment of Type 2 Diabetes<ref name="Bower">PMID:27061187</ref>. As of 2024, it is the only FDA-approved drug for the treatment of Type 2 Diabetes using the AMYR as a target. Recent studies in rodent Alzheimer’s Disease models suggest that pramlintide reduces amyloid-beta plaques, making it a potential therapeutic target for Alzheimer’s Disease<ref name="Grizzanti">PMID:30282360</ref>. | ||
| - | There is limited knowledge about how human amylin binds to the human AMYR, so many studies utilize rat amylin as the peptide for the human AMYR. Rat amylin and pramlintide showcase very similar structures and maintain a high sequence similarity. For instance, the N-term lysine residue is conserved between <scene name='10/1037520/K1/ | + | There is limited knowledge about how human amylin binds to the human AMYR, so many studies utilize rat amylin as the peptide for the human AMYR. Rat amylin and pramlintide showcase very similar structures and maintain a high sequence similarity. For instance, the N-term lysine residue is conserved between <scene name='10/1037520/K1/2'>rat amylin</scene> and <scene name='10/1037516/Pramlintide_k1/1'>pramlintide</scene>, as well as human amylin<ref name="Cao">PMID:35324283</ref>, suggesting that the lysine is integral for binding to AMYR. One of these residue changes is <scene name='10/1038871/Rat_pram_18/1'>R18 in rat amylin to H18 in pramlintide</scene>. Another mutation is <scene name='10/1037520/Rat_pram_19/3'>S19 in rat amylin to K19 in pramlintide</scene>. S19 is highly conserved in most observed variants of amylin, as the <scene name='10/1037520/S19/1'>sidechain of S19 hydrogen bonds to the backbone of P100 of CT</scene><ref name="Cao">PMID:35324283</ref> <ref name="Bower">PMID:27061187</ref>. Other mutations include <scene name='10/1038871/Rat_l23/1'>L23</scene> in rat amylin to <scene name='10/1038871/Pramlintide_f23/1'>F23</scene> in pramlintide and <scene name='10/1038871/Rat_v26/1'>V26</scene> in rat amylin to <scene name='10/1037516/Pramlintide_i26/1'>I26</scene> in pramlintide. While these residues differ, the overall properties of the various residues remain consistent, so many of the same interactions with AMYR are likely still able to form. |
| - | Rat amylin and pramlintide have 3 proline residues, which are absent in human amylin. Human amylin and pramlintide only differ at these 3 residues, with all three being changed to proline in pramlintide. The <scene name='10/1037520/Pramlintide_mutations_labeled/ | + | Rat amylin and pramlintide have 3 proline residues, which are absent in human amylin. Human amylin and pramlintide only differ at these 3 residues, with all three being changed to proline in pramlintide. The <scene name='10/1037520/Pramlintide_mutations_labeled/3'>Ala25Pro, Ser28Pro, and Ser29Pro</scene> mutations break the helical nature present in human amylin, which likely prevents the aggregation of amyloid beta plaques in Alzheimer’s Disease. |
</StructureSection> | </StructureSection> | ||
Current revision
Homo sapiens Amylin3 Receptor, AMYR3
| |||||||||||
References
- ↑ Hay DL, Chen S, Lutz TA, Parkes DG, Roth JD. Amylin: Pharmacology, Physiology, and Clinical Potential. Pharmacol Rev. 2015 Jul;67(3):564-600. PMID:26071095 doi:10.1124/pr.115.010629
- ↑ 2.0 2.1 Grizzanti J, Corrigan R, Casadesus G. Neuroprotective Effects of Amylin Analogues on Alzheimer's Disease Pathogenesis and Cognition. J Alzheimers Dis. 2018;66(1):11-23. PMID:30282360 doi:10.3233/JAD-180433
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 3.9 Cao J, Belousoff MJ, Liang YL, Johnson RM, Josephs TM, Fletcher MM, Christopoulos A, Hay DL, Danev R, Wootten D, Sexton PM. A structural basis for amylin receptor phenotype. Science. 2022 Mar 25;375(6587):eabm9609. PMID:35324283 doi:10.1126/science.abm9609
- ↑ 4.0 4.1 Bower RL, Hay DL. Amylin structure-function relationships and receptor pharmacology: implications for amylin mimetic drug development. Br J Pharmacol. 2016 Jun;173(12):1883-98. PMID:27061187 doi:10.1111/bph.13496
Student Contributors
- Brynn Baker
- Emily Berkman
- Sepp Hall