We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Ben Whiteside
From Proteopedia
(Difference between revisions)
m (Ben Whiteside moved to Sandbox Ben Whiteside: This should be under the protected user domain or Sandbox since using his name as title. And there is already a page https://proteopedia.org/wiki/index.php/User:Ben_Whiteside) |
|||
| (18 intermediate revisions not shown.) | |||
| Line 13: | Line 13: | ||
The amylin peptide contains a <scene name='10/1038819/N_term_disulfide/3'>covalent disulfide linkage</scene> between residues C2 and C7. This disulfide provides stability and rigidity to the helical structure of the peptide, allowing for favorable binding to the extracellular domain (ECD). Notable interactions formed by this disulfide include hydrogen bonds between E294 of the transmembrane domain with K1 of amylin, and both R362 and W361 of the transmembrane domain forming a hydrogen bond with N3 of amylin. | The amylin peptide contains a <scene name='10/1038819/N_term_disulfide/3'>covalent disulfide linkage</scene> between residues C2 and C7. This disulfide provides stability and rigidity to the helical structure of the peptide, allowing for favorable binding to the extracellular domain (ECD). Notable interactions formed by this disulfide include hydrogen bonds between E294 of the transmembrane domain with K1 of amylin, and both R362 and W361 of the transmembrane domain forming a hydrogen bond with N3 of amylin. | ||
====Amidated C-Terminus==== | ====Amidated C-Terminus==== | ||
| - | The <scene name='10/1038819/Amidated_c_term/9'>C-Terminus</scene> of amylin contains an amide group, rather than a carboxylic acid group. This chemical modification allows for more extensive hydrogen bonding to nearby residues, due to the added hydrogen bond donor on the NH2 group. In turn, this allows for favorable hydrogen bonds between S129 of the transmembrane domain and the main chain of Y37 on amylin. This interaction causes a "kink" in the random coil of amylin, displacing Y37 into a hydrophobic pocket, allowing for favorable hydrophobic interactions with W79 of the transmembrane domain. This amidation is thought to be a post-translational modification. | + | The <scene name='10/1038819/Amidated_c_term/9'>C-Terminus</scene> of amylin contains an amide group, rather than a carboxylic acid group. This chemical modification allows for more extensive hydrogen bonding to nearby residues, due to the added hydrogen bond donor on the NH2 group. In turn, this allows for favorable hydrogen bonds between S129 of the transmembrane domain and the main chain of Y37 on amylin. This interaction causes a "kink" in the random coil of amylin, displacing Y37 into a hydrophobic pocket, allowing for favorable hydrophobic interactions with W79 of the transmembrane domain. This amidation is thought to be a post-translational modification. |
==== Bypass Motif ==== | ==== Bypass Motif ==== | ||
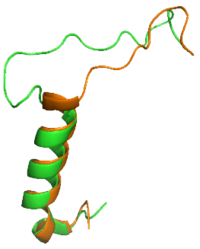
[[Image:SCT_rAmy_Overlay_v2.png|200 px|left|thumb|Figure 1: Overlay of sCT (orange) and rAmy (green)]] | [[Image:SCT_rAmy_Overlay_v2.png|200 px|left|thumb|Figure 1: Overlay of sCT (orange) and rAmy (green)]] | ||
| Line 20: | Line 20: | ||
==Amylin Receptor Binding== | ==Amylin Receptor Binding== | ||
==== Two-Domain Model of Amylin Binding ==== | ==== Two-Domain Model of Amylin Binding ==== | ||
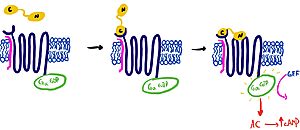
| - | It is hypothesized that amylin binds to the receptor via a two-domain model. The model suggests a series of steps for how amylin binds. First, the c-terminus of amylin binds to the n terminus of the extracellular domain of the receptor. This binding factors the alignment of amylin's n-terminus to the primary GPCR binding site. Once both the c-terminus and n-terminus of amylin are bound, the receptor becomes activated [[Image:Domain_drawingnew.jpg|300px|left|thumb|Figure 2: The Two Domain Model]] | + | It is hypothesized that amylin binds to the receptor via a two-domain model. The model suggests a series of steps for how amylin binds. First, the c-terminus of amylin binds to the n terminus of the extracellular domain of the receptor. This binding factors the alignment of amylin's n-terminus to the primary GPCR binding site. Once both the c-terminus and n-terminus of amylin are bound, the receptor becomes activated. [[Image:Domain_drawingnew.jpg|300px|left|thumb|Figure 2: The Two Domain Model]] |
====RAMP-CTR Interface==== | ====RAMP-CTR Interface==== | ||
<scene name='10/1038828/Ramp_ctr_interface/9'>RAMP CTR Interface </scene> is a key interaction that stabilizes the protein complex and positions the receptor to favorably bind to amylin. The RAMP-CTR interface extends into the plasma membrane, providing additional non-covalent bonding between the protein complex and the cell membrane. | <scene name='10/1038828/Ramp_ctr_interface/9'>RAMP CTR Interface </scene> is a key interaction that stabilizes the protein complex and positions the receptor to favorably bind to amylin. The RAMP-CTR interface extends into the plasma membrane, providing additional non-covalent bonding between the protein complex and the cell membrane. | ||
Current revision
AMYR
| |||||||||||
Student Contributors
Andrew Helmerich, Mathias Vander Eide, Ben Whiteside