We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Mandy Bechman/Sandbox 1
From Proteopedia
< User:Mandy Bechman(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 33: | Line 33: | ||
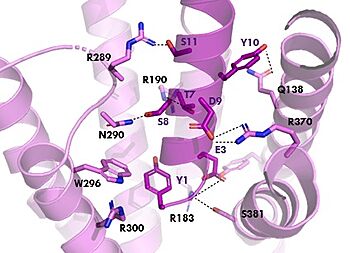
[[Image:TZ_hydrogen_bonds.jpg|350 px|right|thumb|GIPR (pink) and Tirzepatide (dark pink) residue interactions]] | [[Image:TZ_hydrogen_bonds.jpg|350 px|right|thumb|GIPR (pink) and Tirzepatide (dark pink) residue interactions]] | ||
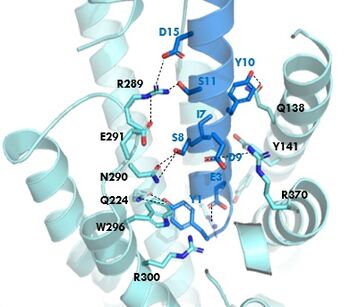
The <scene name='10/1038815/Tirzepatide_active_site/4'>binding site</scene> with Tirzepatide is the same as GIP with the N-term binding to the transmembrane domain and activating cellular signaling.The <scene name='10/1038815/Tirzepatide_residues/7'>residues</scene> are fairly similar just in a different conformations, which is allowing for more hydrogen bonding. In Tirzepatide, the most noticeable conformational change is the Tyrosine 1 (Y1) residue. It is now facing up towards Arginine 190 (R190) where they can now form a new hydrogen bond that was not present in GIP. The more hydrogen bonding there is between molecules, the stronger the binding between the two molecules will be. This can lead to the conclusion that GIPR has a higher binding affinity for Tirzepatide than GIP itself<ref name="Sun"/>. | The <scene name='10/1038815/Tirzepatide_active_site/4'>binding site</scene> with Tirzepatide is the same as GIP with the N-term binding to the transmembrane domain and activating cellular signaling.The <scene name='10/1038815/Tirzepatide_residues/7'>residues</scene> are fairly similar just in a different conformations, which is allowing for more hydrogen bonding. In Tirzepatide, the most noticeable conformational change is the Tyrosine 1 (Y1) residue. It is now facing up towards Arginine 190 (R190) where they can now form a new hydrogen bond that was not present in GIP. The more hydrogen bonding there is between molecules, the stronger the binding between the two molecules will be. This can lead to the conclusion that GIPR has a higher binding affinity for Tirzepatide than GIP itself<ref name="Sun"/>. | ||
| - | + | === Isoleucine vs. Threonine === | |
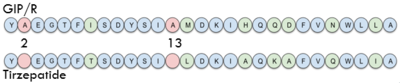
| - | + | In GIP there is an Isoleucine present <scene name='10/1038815/Gip_ile7/3'>(I7)</scene>. Isoleucine is a branched residue that is very hydrophobic. This residue is also present in the N-term where the important interactions are occuring. In Tirzepatide, this seventh residue has been changed to a Threonine residue <scene name='10/1038815/Tirzepatide_thr7/3'>(T7)</scene>. Threonine is also a branched residue, but it contains a hydroxyl group making it hydrophilic, allowing it to make hydrogen bonds. Threonine 7 (T7) is also making a hydrogen bone with the Arginine 190 (R190) residue. This extra hydrogen bond in Tirzepatide is also responsible for a higher binding affinity for the drug than GIP itself. | |
| + | ===AIB Mutation=== | ||
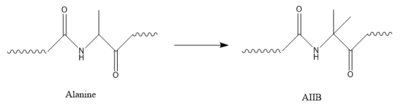
[[Image:AlatoAIB.png|400 px|left|thumb|Structure of alanine converted to AIB.]] | [[Image:AlatoAIB.png|400 px|left|thumb|Structure of alanine converted to AIB.]] | ||
| + | In GIP, residue 2 is an <scene name='10/1038867/Alanine_2_-_gip/3'>alanine</scene> but in Tirzepatide, the residue changes to <scene name='10/1038867/Tirzepatide_-_aib/2'>AIB</scene>. Residue 2 is where DPP4 recognizes GIP in order to cleave and inactivate the hormone. When the residue is changed, it is no longer recognized by DPP4 because AIB is a noncoding residue, resulting in continuous signaling to the cell. | ||
| - | |||
| - | == Relevance == | ||
| - | In GIP, the residue 2 is an <scene name='10/1038867/Alanine_2_-_gip/3'>alanine</scene> but in Tirzepatide, the residue changes to <scene name='10/1038867/Tirzepatide_-_aib/2'>AIB</scene> | ||
| - | |||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
Current revision
H. sapiens Glucose-dependent Insulinotropic Polypeptide
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Mayendraraj A, Rosenkilde MM, Gasbjerg LS. GLP-1 and GIP receptor signaling in beta cells interactions and co-stimulation. Peptides. 2022 May;151:170749. PMID:35065096 doi:10.1016/j.peptides.2022.170749
- ↑ 2.0 2.1 2.2 2.3 Seino Y, Fukushima M, Yabe D. GIP and GLP-1, the two incretin hormones: Similarities and differences. J Diabetes Investig. 2010 Apr 22;1(1-2):8-23. PMID:24843404 doi:10.1111/j.2040-1124.2010.00022.x
- ↑ 3.0 3.1 3.2 Sun B, Willard FS, Feng D, Alsina-Fernandez J, Chen Q, Vieth M, Ho JD, Showalter AD, Stutsman C, Ding L, Suter TM, Dunbar JD, Carpenter JW, Mohammed FA, Aihara E, Brown RA, Bueno AB, Emmerson PJ, Moyers JS, Kobilka TS, Coghlan MP, Kobilka BK, Sloop KW. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2116506119. PMID:35333651 doi:10.1073/pnas.2116506119
Student Contributors
Mandy M. Bechman Chloe A. Tucker