We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Isabel Kluszynski/Sandbox 1
From Proteopedia
< User:Isabel Kluszynski(Difference between revisions)
| (7 intermediate revisions not shown.) | |||
| Line 14: | Line 14: | ||
===Binding Interactions of GLP-1=== | ===Binding Interactions of GLP-1=== | ||
| - | GLP-1 engages in several <scene name='10/1037487/Nterm_residues/23'>binding interactions</scene> with the GLP-1R | + | GLP-1 engages in several <scene name='10/1037487/Nterm_residues/23'>binding interactions</scene> with the GLP-1R. The N-terminus of GLP-1 binds within the transmembrane region of GLP-1R, and the C-terminus of GLP-1 binds to the N-terminus of GLP-1R. Upon GLP-1 binding, a signal cascade is triggered beginning with an elevation in cAMP levels. Increased intracellular [https://en.wikipedia.org/wiki/Cyclic_adenosine_monophosphate cAMP] levels activate [https://en.wikipedia.org/wiki/Protein_kinase_A PKA] and EPAC2 leading to several downstream signaling effects including increased ATP production, increased insulin production and secretion, and membrane depolarization. <ref name="Mayendraraj"/> Beginning at the N-terminus of GLP-1, <scene name='10/1037487/Nterm_residues/26'>GLP-1 E9</scene> forms a salt bridge with GLP-1R R190 and a hydrogen bond with GLP-1R Y152. Next, <scene name='10/1037487/Nterm_residues/22'>GLP-1 T13</scene> hydrogen bonds to GLP-1R K197. Towards the middle of the peptide chain, <scene name='10/1037487/Nterm_residues/25'>GLP-1 S17</scene> hydrogen bonds with GLP-1R R299 and GLP-1 E21 hydrogen bonds with GLP-1R Y205. At the <scene name='10/1037487/Cterm_glp/9'>C-terminus</scene> of GLP-1, <scene name='10/1037487/Cterm_glp/10'>GLP-1 E27</scene> hydrogen bonds with GLP-1R Q210. Right above this, there is a pi stacking interaction between GLP-1 F28 and W31 with GLP-1R W214. <ref name="Zhao">PMID:35217653</ref> |
===Binding Interactions of Tirzepatide=== | ===Binding Interactions of Tirzepatide=== | ||
| Line 24: | Line 24: | ||
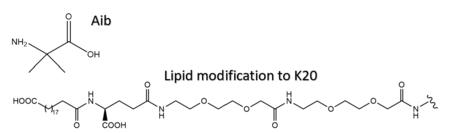
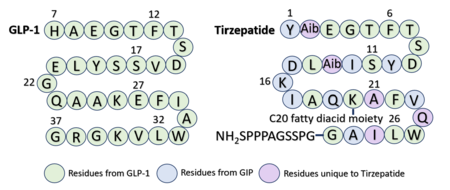
Several residues have been selectively and intentionally modified during Tirzepatide drug design. <scene name='10/1037487/Interesting_tirz_modifications/4'>Aib</scene>, aminoisobutyric acid, is located at positions 2 and 13 with the purpose of preventing [https://en.wikipedia.org/wiki/Dipeptidyl_peptidase-4#:~:text=Dipeptidyl%20peptidase-4%20%28DPP4%20or%20DPPIV%29%2C%20also%20known%20as,DPP4%20is%20related%20to%20FAP%2C%20DPP8%2C%20and%20DPP9. DPP-4] cleavage and inactivation. K20 of Tirzepatide has a lipid modification, specifically a C20 fatty diacid moiety, that serves to enhance binding to the protein carrier albumin and increase the half-life of the drug in the body. <ref name="Sun"/> | Several residues have been selectively and intentionally modified during Tirzepatide drug design. <scene name='10/1037487/Interesting_tirz_modifications/4'>Aib</scene>, aminoisobutyric acid, is located at positions 2 and 13 with the purpose of preventing [https://en.wikipedia.org/wiki/Dipeptidyl_peptidase-4#:~:text=Dipeptidyl%20peptidase-4%20%28DPP4%20or%20DPPIV%29%2C%20also%20known%20as,DPP4%20is%20related%20to%20FAP%2C%20DPP8%2C%20and%20DPP9. DPP-4] cleavage and inactivation. K20 of Tirzepatide has a lipid modification, specifically a C20 fatty diacid moiety, that serves to enhance binding to the protein carrier albumin and increase the half-life of the drug in the body. <ref name="Sun"/> | ||
| - | Similarly to GLP-1 forming several | + | Similarly to GLP-1 forming several ligand binding interactions with GLP-1R, Tirzepatide also forms many key <scene name='10/1037487/Tirzepatidebonding/8'>stabilizing interactions</scene> with GLP-1R. Beginning at the N-terminus of Tirzepatide (Tirz), <scene name='10/1037487/Tirzepatidebonding/9'>Tirz E3</scene> forms a salt bridge with GLP-1R R190 and a hydrogen bond with GLP-1R Y152. Additionally, Tirz T7 hydrogen bonds with GLP-1R K197. These interactions are nearly identical to the interactions GLP-1 E9 and T13 make with the receptor. Towards the middle of the peptide, <scene name='10/1037487/Tirzepatidebonding/6'>Tirz D15</scene> forms a hydrogen bond with Y205, yet another similar interaction to GLP-1 binding. <ref name="Zhao"/> Looking at the sequence comparison of GLP-1 and Tirzepatide, GLP-1 E21 is located at the same position as Tirz D15. |
===Comparison of Binding Interactions=== | ===Comparison of Binding Interactions=== | ||
| Line 32: | Line 32: | ||
Through an <scene name='10/1037490/Overlay/7'>overlay</scene>, it is visible that GLP-1 and Tirzepatide bind similarly but not identically to the receptor. Some interactions seen in GLP-1 bound to GLP-1R are preserved with Tirzepatide, while others are altered. For example, the first residue of GLP-1 is a <scene name='10/1037487/Gvt_binding/7'>histidine</scene>, while the first residue of Tirzepatide is a <scene name='10/1037487/Gvt_binding/9'>tyrosine</scene>. <ref name="Sun"/> The identity of this first residue can either favor or prevent specific GLP-1R interactions. With GLP-1 H7, <scene name='10/1037487/Gvt_binding/11'>GLP-1R R310</scene> and E373 are in close enough proximity to form a salt bridge. Additionally, GLP-1R W306 and D372 are positioned ideally for hydrogen bond formation. However, with Tirzepatide bound, Tirz Y1 causes greater steric clashing, pushing the <scene name='10/1037487/Gvt_binding/10'>GLP-1R R310</scene> and other residues further apart so they are unable to interact. <ref name="Sun"/> | Through an <scene name='10/1037490/Overlay/7'>overlay</scene>, it is visible that GLP-1 and Tirzepatide bind similarly but not identically to the receptor. Some interactions seen in GLP-1 bound to GLP-1R are preserved with Tirzepatide, while others are altered. For example, the first residue of GLP-1 is a <scene name='10/1037487/Gvt_binding/7'>histidine</scene>, while the first residue of Tirzepatide is a <scene name='10/1037487/Gvt_binding/9'>tyrosine</scene>. <ref name="Sun"/> The identity of this first residue can either favor or prevent specific GLP-1R interactions. With GLP-1 H7, <scene name='10/1037487/Gvt_binding/11'>GLP-1R R310</scene> and E373 are in close enough proximity to form a salt bridge. Additionally, GLP-1R W306 and D372 are positioned ideally for hydrogen bond formation. However, with Tirzepatide bound, Tirz Y1 causes greater steric clashing, pushing the <scene name='10/1037487/Gvt_binding/10'>GLP-1R R310</scene> and other residues further apart so they are unable to interact. <ref name="Sun"/> | ||
| - | Another example of a difference in receptor | + | Another example of a difference in receptor conformation can be seen with GLP-1R R299. With GLP-1 bound, <scene name='10/1037487/Glp1_comparison/3'>GLP-1R R299</scene> faces the peptide and is able to hydrogen bond with either GLP-1 S17 or E21. When Tirzepatide is bound, <scene name='10/1037487/Tirzepatide/7'>GLP-1R R299</scene> flips away from the peptide and can no longer hydrogen bond with any Tirzepatide residues. <ref name="Sun"/> Additionally, differential binding of GLP-1 and Tirzepatide modifies how the transmembrane domain interacts with the G-alpha subunit to initiate a signal cascade. For example, when GLP-1 is bound to the receptor, <scene name='10/1037487/G_alpha_glp/2'>GLP-1R F257</scene> is able to participate in a pi stacking interaction with G-alpha F376. When Tirzepatide is bound, <scene name='10/1037487/G_alpha_tirz/3'>GLP-1R F257</scene> is oriented facing away from the G-alpha subunit. <ref name="Zhao"/> |
Referencing the sequence alignment, GLP-1 F28 and W31 are in the same position as Tirzepatide F22 and W25, indicating that the phenylalanine and tryptophan residues are conserved among the two sequences. As discussed previously in the GLP-1 binding interactions subsection, <scene name='10/1037487/Cterm_glp/11'>GLP-1 F28</scene> and W31 participate in a pi stacking interaction with GLP-1R W214. Similarly, <scene name='10/1037487/Tirzepatide/8'>Tirz F22</scene> and W25 are also able to interact aromatically with GLP-1R W214. | Referencing the sequence alignment, GLP-1 F28 and W31 are in the same position as Tirzepatide F22 and W25, indicating that the phenylalanine and tryptophan residues are conserved among the two sequences. As discussed previously in the GLP-1 binding interactions subsection, <scene name='10/1037487/Cterm_glp/11'>GLP-1 F28</scene> and W31 participate in a pi stacking interaction with GLP-1R W214. Similarly, <scene name='10/1037487/Tirzepatide/8'>Tirz F22</scene> and W25 are also able to interact aromatically with GLP-1R W214. | ||
| Line 38: | Line 38: | ||
==Medical Relevance== | ==Medical Relevance== | ||
| - | There have been several drugs designed to target the GLP-1 receptor. In 2014, Trulicity (dulaglutide) was FDA approved for weekly injections as treatment for Type 2 Diabetes. In 2017, Ozempic (semaglutide) was FDA approved as both a weekly injection or a daily oral pill to regulate blood glucose. The GLP-1R/GIP-R coagonist Mounjaro (tirzepatide), FDA approved in 2022, has shown a significant superiority over GLP-1 agonists alone as both a treatment for Type 2 Diabetes and as a weight loss drug. <ref name="Nauck">PMID:33068776</ref> Promising initial clinical trial results are now being demonstrated for retatrutide: a GLP-1R/GIP-R/glucagon (GCG) receptor triple hormone agonist. Data from phase 2 clinical trials reveal an average of 24.2% weight loss over the course of 48 weeks, suggesting retatrutide as a viable treatment for obesity. <ref name="Naeem">PMID:38323122</ref> | + | There have been several drugs designed to target the GLP-1 receptor. In 2014, Trulicity (dulaglutide) was FDA approved for weekly injections as treatment for Type 2 Diabetes. In 2017, Ozempic (semaglutide) was FDA approved as both a weekly injection or a daily oral pill to regulate blood glucose. The GLP-1R/GIP-R coagonist Mounjaro (tirzepatide), FDA approved in 2022, has shown a significant superiority over GLP-1 agonists alone as both a treatment for Type 2 Diabetes and as a weight loss drug. <ref name="Nauck">PMID:33068776</ref> Promising initial clinical trial results are now being demonstrated for retatrutide: a GLP-1R/GIP-R/glucagon (GCG) receptor triple hormone agonist. Data from phase 2 clinical trials reveal an average of 24.2% weight loss over the course of 48 weeks, suggesting retatrutide as a viable future treatment for obesity. <ref name="Naeem">PMID:38323122</ref> |
</StructureSection> | </StructureSection> | ||
Current revision
Glucagon-like peptide-1 receptor (GLP-1R) Homo sapiens
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Mayendraraj A, Rosenkilde MM, Gasbjerg LS. GLP-1 and GIP receptor signaling in beta cells interactions and co-stimulation. Peptides. 2022 May;151:170749. PMID:35065096 doi:10.1016/j.peptides.2022.170749
- ↑ 2.0 2.1 Seino Y, Fukushima M, Yabe D. GIP and GLP-1, the two incretin hormones: Similarities and differences. J Diabetes Investig. 2010 Apr 22;1(1-2):8-23. PMID:24843404 doi:10.1111/j.2040-1124.2010.00022.x
- ↑ Zhang X, Belousoff MJ, Zhao P, Kooistra AJ, Truong TT, Ang SY, Underwood CR, Egebjerg T, Šenel P, Stewart GD, Liang YL, Glukhova A, Venugopal H, Christopoulos A, Furness SGB, Miller LJ, Reedtz-Runge S, Langmead CJ, Gloriam DE, Danev R, Sexton PM, Wootten D. Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists. Mol Cell. 2020 Nov 5;80(3):485-500.e7. PMID:33027691 doi:10.1016/j.molcel.2020.09.020
- ↑ 4.0 4.1 4.2 Zhao F, Zhou Q, Cong Z, Hang K, Zou X, Zhang C, Chen Y, Dai A, Liang A, Ming Q, Wang M, Chen LN, Xu P, Chang R, Feng W, Xia T, Zhang Y, Wu B, Yang D, Zhao L, Xu HE, Wang MW. Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors. Nat Commun. 2022 Feb 25;13(1):1057. PMID:35217653 doi:10.1038/s41467-022-28683-0
- ↑ 5.0 5.1 5.2 5.3 5.4 Sun B, Willard FS, Feng D, Alsina-Fernandez J, Chen Q, Vieth M, Ho JD, Showalter AD, Stutsman C, Ding L, Suter TM, Dunbar JD, Carpenter JW, Mohammed FA, Aihara E, Brown RA, Bueno AB, Emmerson PJ, Moyers JS, Kobilka TS, Coghlan MP, Kobilka BK, Sloop KW. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2116506119. PMID:35333651 doi:10.1073/pnas.2116506119
- ↑ Nauck MA, Quast DR, Wefers J, Meier JJ. GLP-1 receptor agonists in the treatment of type 2 diabetes Mol Metab. 2021 Apr;46:101102. PMID:33068776 doi:10.1016/j.molmet.2020.101102
- ↑ Naeem M, Imran L, Banatwala UESS. Unleashing the power of retatrutide: A possible triumph over obesity and overweight: A correspondence. Health Sci Rep. 2024 Feb 5;7(2):e1864. PMID:38323122 doi:10.1002/hsr2.1864
Student Contributors
- Isabel Kluszynski
- Makenna Marcinek