We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox324
From Proteopedia
(Difference between revisions)
| (8 intermediate revisions not shown.) | |||
| Line 46: | Line 46: | ||
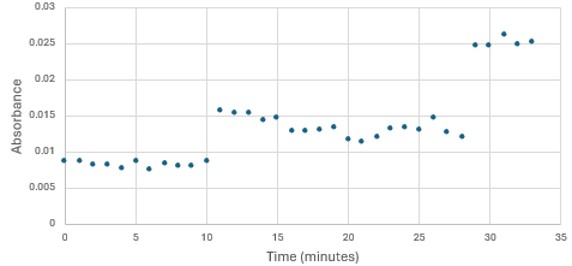
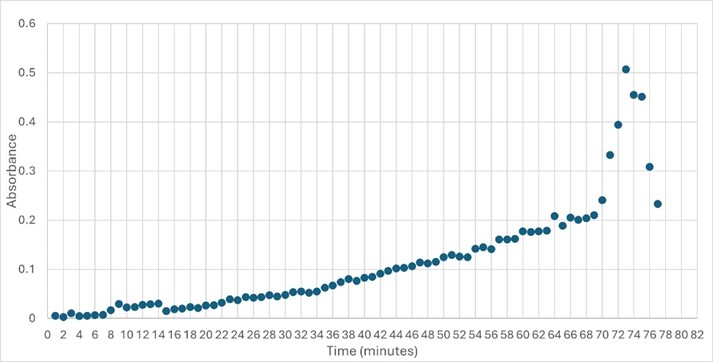
'''Figure 5: ''' Trial 2 of 4DIU enzymatic activity measured over time via change in absorbance at 405nm in pH 6 buffer | '''Figure 5: ''' Trial 2 of 4DIU enzymatic activity measured over time via change in absorbance at 405nm in pH 6 buffer | ||
| - | == Structural highlights of 4DIU == | + | == Structural highlights of 4DIU == |
| - | + | ||
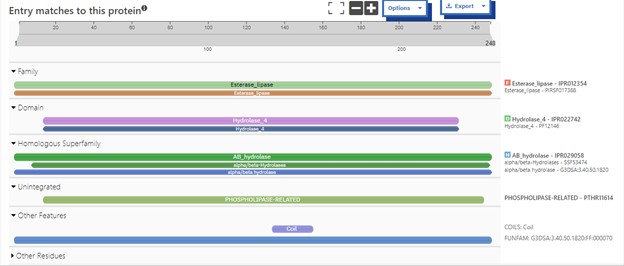
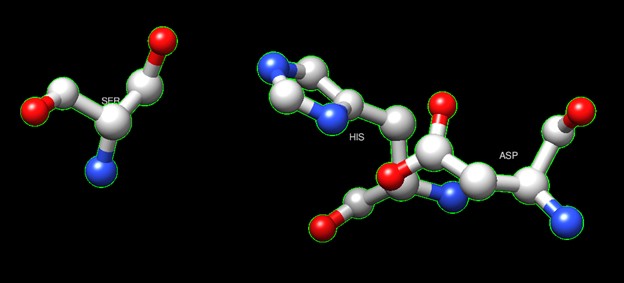
One of the distinctive structural characteristics of this protein is the alpha/beta-hydrolase (ABH) fold. Evidence for this fold consists of its structure (an open β-sheet surrounded by α-helices) and the presence of what is known as the "catalytic triad" (consisting of a nucleophile, an acid, and histidine)<ref>Holmquist, M. Alpha Beta-Hydrolase Fold Enzymes Structures, Functions and Mechanisms. Current Protein and Peptide Science 2000, 1 (2), 209–235. https://doi.org/10.2174/1389203003381405.</ref>. The presence of the active site residues His A 222, Asp A 192, and Ser A 93, as determined by SPRITE, Chimera, etc., confirms the presence of this catalytic triad. | One of the distinctive structural characteristics of this protein is the alpha/beta-hydrolase (ABH) fold. Evidence for this fold consists of its structure (an open β-sheet surrounded by α-helices) and the presence of what is known as the "catalytic triad" (consisting of a nucleophile, an acid, and histidine)<ref>Holmquist, M. Alpha Beta-Hydrolase Fold Enzymes Structures, Functions and Mechanisms. Current Protein and Peptide Science 2000, 1 (2), 209–235. https://doi.org/10.2174/1389203003381405.</ref>. The presence of the active site residues His A 222, Asp A 192, and Ser A 93, as determined by SPRITE, Chimera, etc., confirms the presence of this catalytic triad. | ||
| - | [ | + | [[Image:Activesite.jpeg]] |
| - | Another distinctive feature of this protein that demonstrates its identity as an esterase is a coil that correlates to the bioinformatic predictions of Chimera, BLAST, Dali, and Sprite. | + | |
| + | '''Figure 6''':Catalytic triad of protein 4DIU | ||
| + | |||
| + | Another distinctive feature of this protein that demonstrates its identity as an esterase is a coil that correlates to the bioinformatic predictions of Chimera, BLAST, Dali, and Sprite.<ref>PMID:21638687</ref> | ||
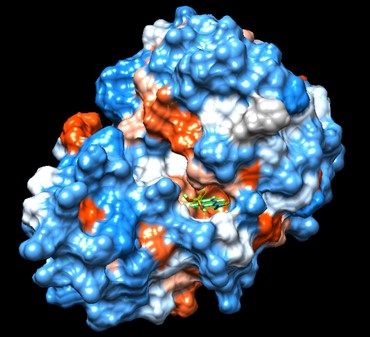
Swiss Dock and Chimera predicted that this protein would have binding sites with an affinity to substrates such as acetate, butyrate, phosphate, proline, decanoate, dodecanoate, etc. Wet lab experiments still must be conducted in order to confirm these predictions. | Swiss Dock and Chimera predicted that this protein would have binding sites with an affinity to substrates such as acetate, butyrate, phosphate, proline, decanoate, dodecanoate, etc. Wet lab experiments still must be conducted in order to confirm these predictions. | ||
| + | [[Image:docking.jpg]] | ||
| + | |||
| + | '''Figure 7''': Docking of cluster 1.2 of proline with protein 4DIU | ||
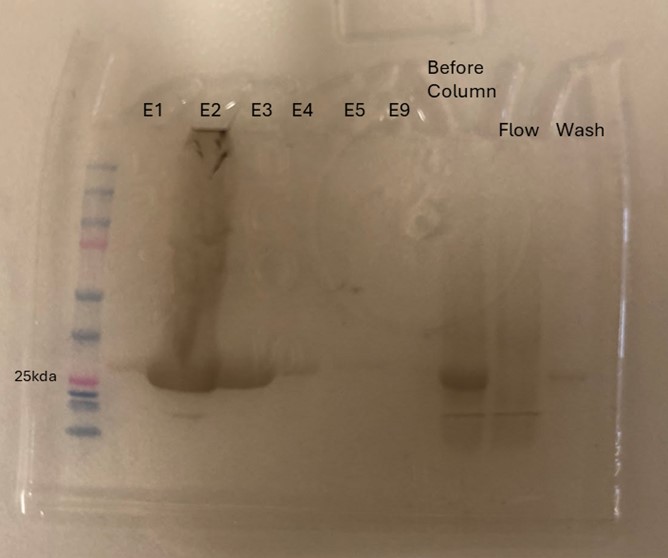
| + | Protein 4DIU was calculated to have a molecular weight of 27.28 kDa. This was found by taking 248*110, 248 is the number of amino acids and 110 is the average weight in daltons of an amino acid. This size was confirmed by running an SDS-Page gel of multiple elutions isolated from a column as well as samples from purification steps along the way. | ||
| + | [[Image:sGel.jpeg]] | ||
| + | |||
| + | '''Figure 8''': SDS-Page gel confirming size and presence of protein 4DIU | ||
== Conclusions == | == Conclusions == | ||
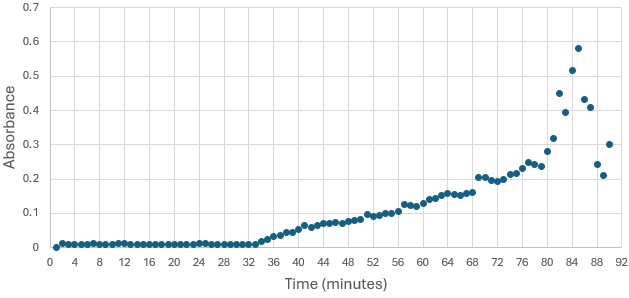
| - | Overall, the research question was answered and the hypothesis was correct. Protein 4DIU has been identified as belonging to the esterase family and has hydrolytic activity. It appears that the optimal pH for this enzyme is 6 and it is active with p-nitrophenyl acetate. These results are fairly accurate and precise as two trials were run at a pH of 6 and the same trend was seen. Again, from this data, it is believed that protein 4DIU would be active in | + | Overall, the research question was answered and the hypothesis was correct. Protein 4DIU has been identified as belonging to the esterase family and has hydrolytic activity. It appears that the optimal pH for this enzyme is 6 and it is active with p-nitrophenyl acetate. These results are fairly accurate and precise as two trials were run at a pH of 6 and the same trend was seen. Again, from this data, it is believed that protein 4DIU would be active in liver and skin cells. In future experiments, it would be beneficial to test the activity at lower pHs such as 3, 4, 5, and 7. This could rule out certain parts of the body if the lower pH causes inactivation. Additionally, testing some of the substrates that were modeled to work to see if the bioinformatic data is trustworthy. |
</StructureSection> | </StructureSection> | ||
Current revision
| |||||||||||
References
- ↑ Fukami, T.; Yokoi, T. The Emerging Role of Human Esterases. Drug Metabolism and Pharmacokinetics 2012, 27 (5), 466–477. https://doi.org/10.2133/dmpk.dmpk-12-rv-042.
- ↑ Tokudome, Y.; Katayanagi, M.; Hashimoto, F. Esterase Activity and Intracellular Localization in Reconstructed Human Epidermal Cultured Skin Models. Annals of Dermatology 2015, 27 (3), 269. https://doi.org/10.5021/ad.2015.27.3.269.
- ↑ Williams, F. M. Clinical Significance of Esterases in Man. Clinical pharmacokinetics 1985, 10 (5), 392–403. https://doi.org/10.2165/00003088-198510050-00002.
- ↑ Zhang, S.; Sun, W.; Xu, L.; Zheng, X.; Chu, X.; Tian, J.; Wu, N.; Fan, Y. Identification of the Para-Nitrophenol Catabolic Pathway, and Characterization of Three Enzymes Involved in the Hydroquinone Pathway, in Pseudomonas Sp. 1-7. BMC Microbiology 2012, 12 (1). https://doi.org/10.1186/1471-2180-12-27.
- ↑ Vázquez-Mayorga, E.; Díaz-Sánchez, Á.; Dagda, R.; Domínguez-Solís, C.; Dagda, R.; Coronado-Ramírez, C.; Martínez-Martínez, A. Novel Redox-Dependent Esterase Activity (EC 3.1.1.2) for DJ-1: Implications for Parkinson’s Disease. International Journal of Molecular Sciences 2016, 17 (8), 1346. https://doi.org/10.3390/ijms17081346.
- ↑ Holmquist, M. Alpha Beta-Hydrolase Fold Enzymes Structures, Functions and Mechanisms. Current Protein and Peptide Science 2000, 1 (2), 209–235. https://doi.org/10.2174/1389203003381405.
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644