User:Karisma Moll/Sandbox 1
From Proteopedia
< User:Karisma Moll(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 23: | Line 23: | ||
The <scene name='10/1037489/Cystine_rich_region/2'>cystine rich region</scene> contains 6 cystine residues (C385, C394, C444, C447, C454, C472) that make <scene name='10/1037489/Disulfide_bonds/2'>three disulfide bonds</scene> that play a critical role in the tertiary structure and therefore function of the DPPIV enzyme. <ref name="Dobers">PMID:10931192</ref> | The <scene name='10/1037489/Cystine_rich_region/2'>cystine rich region</scene> contains 6 cystine residues (C385, C394, C444, C447, C454, C472) that make <scene name='10/1037489/Disulfide_bonds/2'>three disulfide bonds</scene> that play a critical role in the tertiary structure and therefore function of the DPPIV enzyme. <ref name="Dobers">PMID:10931192</ref> | ||
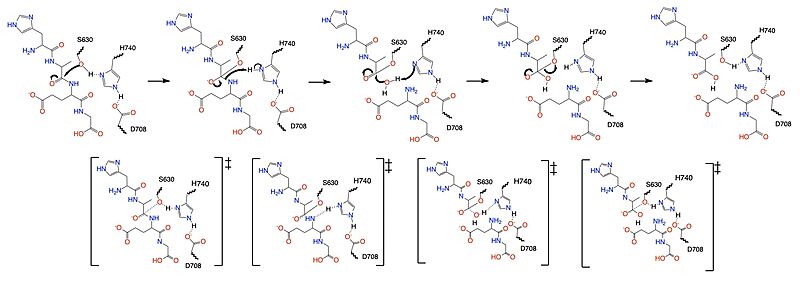
| - | The {{font color|mediumseagreen|catalytic domain}} is where DPP-IV substrates are cleaved at the penultimate point or where inhibitors bind to prevent DPP-IV enzymatic activity.The <scene name='10/1037489/Catalytic_domain/2'>catalytic domain</scene> includes the {{font color|springgreen|S1 binding pocket}} and {{font color|teal|S2 binding pocket}}. The {{font color|springgreen|S1 binding pocket}} contains hydrophobic residues (W547, S630, Y631, V656, W659, Y662, Y666, N710, V711 and H740) that interact with either the substrate or inhibitor to keep it within the catalytic domain. The {{font color|teal|S2 binding pocket}} contains residues (E205, E206, Y662, S209, R358 and P357) that create hydrogen bonds with either substrate or inhibitor to additionally keep it in place to be cleaved by the catalytic triad. <ref name="Metzler">PMID:18227430</ref> Within these <scene name='10/1037489/Bp_w_surface_new/2'>binding pockets</scene> is the <scene name='10/1037489/Catalytic_triad/ | + | The {{font color|mediumseagreen|catalytic domain}} is where DPP-IV substrates are cleaved at the penultimate point or where inhibitors bind to prevent DPP-IV enzymatic activity.The <scene name='10/1037489/Catalytic_domain/2'>catalytic domain</scene> includes the {{font color|springgreen|S1 binding pocket}} and {{font color|teal|S2 binding pocket}}. The {{font color|springgreen|S1 binding pocket}} contains hydrophobic residues (W547, S630, Y631, V656, W659, Y662, Y666, N710, V711 and H740) that interact with either the substrate or inhibitor to keep it within the catalytic domain. The {{font color|teal|S2 binding pocket}} contains residues (E205, E206, Y662, S209, R358 and P357) that create hydrogen bonds with either substrate or inhibitor to additionally keep it in place to be cleaved by the catalytic triad. <ref name="Metzler">PMID:18227430</ref> Within these <scene name='10/1037489/Bp_w_surface_new/2'>binding pockets</scene> is the <scene name='10/1037489/Catalytic_triad/10'>catalytic triad</scene>, assisted by the oxyanion hole (Y631). <ref name="Kim">PMID:30103438</ref> |
=== Mechanism === | === Mechanism === | ||
| Line 40: | Line 40: | ||
Diabetes mellitus is a metabolic disorder, characterized by hyperglycemia, caused by irregularity in insulin secretion, insulin action, or a combination of both. [https://en.wikipedia.org/wiki/Type_1_diabetes Type 1 diabetes mellitus] (T1DM) is characterized by the autoimmune destruction of pancreatic beta-cells, leading to diabetic ketoacidosis. [https://en.wikipedia.org/wiki/Type_2_diabetes Type 2 diabetes mellitus] (T2DM) is characterized by insulin resistance and subsequent deficiency. <ref name="Banday">PMID:33437689</ref> | Diabetes mellitus is a metabolic disorder, characterized by hyperglycemia, caused by irregularity in insulin secretion, insulin action, or a combination of both. [https://en.wikipedia.org/wiki/Type_1_diabetes Type 1 diabetes mellitus] (T1DM) is characterized by the autoimmune destruction of pancreatic beta-cells, leading to diabetic ketoacidosis. [https://en.wikipedia.org/wiki/Type_2_diabetes Type 2 diabetes mellitus] (T2DM) is characterized by insulin resistance and subsequent deficiency. <ref name="Banday">PMID:33437689</ref> | ||
| - | <scene name='10/1037489/Glp1_peptide/2'>Glucagon-like Peptide-1</scene> (GLP-1) is a 30 amino acid hormone secreted into the gut by intestinal epithelial L-cells. GLP-1 is secreted in response to meal intake and is responsible for stimulating insulin production. GLP-1 binding to the GLP-1R located on the membranes of pancreatic cells results in the upregulation of insulin production and secretion which directly results in the lowering of blood glucose levels. <ref name="Holst">PMID:17928588</ref> | + | <scene name='10/1037489/Glp1_peptide/2'>Glucagon-like Peptide-1</scene> (GLP-1) is a 30 amino acid hormone secreted into the gut by intestinal epithelial L-cells. GLP-1 is secreted in response to meal intake and is responsible for stimulating insulin production. GLP-1 binding to the <scene name='10/1037491/Glp-1r/5'>GLP-1R</scene> located on the membranes of pancreatic cells results in the upregulation of insulin production and secretion which directly results in the lowering of blood glucose levels. <ref name="Holst">PMID:17928588</ref> |
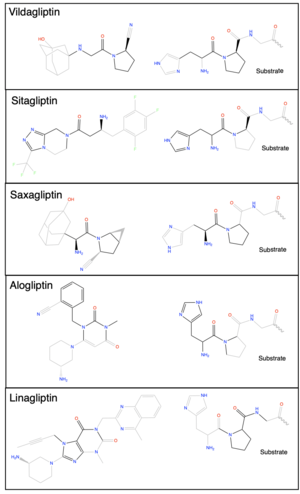
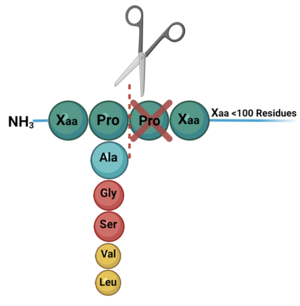
DPP-IV binds and degrades GLP-1 <scene name='10/1037489/Glp1_peptide__scissile_bond/3'>(cleaving at the penultimate position)</scene> resulting in heightened blood glucose levels. Insufficient GLP-1 production or signaling in response to meal intake is clinically associated with T2DM and morbidity. Since GLP-1 is a potent regulator of blood glucose levels, and DPP-IV antagonistically regulates GLP-1, this makes DPP-IV inhibitors an excellent candidate for pharmacological therapeutics for T2DM. <ref name="Gilbert">PMID:32308645</ref> | DPP-IV binds and degrades GLP-1 <scene name='10/1037489/Glp1_peptide__scissile_bond/3'>(cleaving at the penultimate position)</scene> resulting in heightened blood glucose levels. Insufficient GLP-1 production or signaling in response to meal intake is clinically associated with T2DM and morbidity. Since GLP-1 is a potent regulator of blood glucose levels, and DPP-IV antagonistically regulates GLP-1, this makes DPP-IV inhibitors an excellent candidate for pharmacological therapeutics for T2DM. <ref name="Gilbert">PMID:32308645</ref> | ||
| - | Recent studies are demonstrating connections between regulation of [https://en.wikipedia.org/wiki/Caveolin_1 Caveolin-1] (Cav-1) and subsequent GLP-1 action. Cav-1, the principal protein of caveolae, is an integral membrane protein that co-localizes with GLP-1R to regulate receptor trafficking and control the assembly of signaling molecules. Direct interaction of Cav-1 with [https://en.wikipedia.org/wiki/Glucagon-like_peptide-1_receptor GLP-1R] is required for trafficking and regulation. This interaction occurs with the binding motif in the second intracellular loop of GLP-1R. Two residues in particular, | + | Recent studies are demonstrating connections between regulation of [https://en.wikipedia.org/wiki/Caveolin_1 Caveolin-1] (Cav-1) and subsequent GLP-1 action. <scene name='10/1037493/Cav-1_overview/3'>Cav-1</scene>, the principal protein of caveolae, is an integral membrane protein that co-localizes with GLP-1R to regulate receptor trafficking and control the assembly of signaling molecules. Direct interaction of Cav-1 with [https://en.wikipedia.org/wiki/Glucagon-like_peptide-1_receptor GLP-1R] is required for trafficking and regulation. This interaction occurs with the binding motif in the second intracellular loop of GLP-1R. Two residues in particular, <scene name='10/1037491/Glp-1r/1'>Y250 and Y252</scene>, were proved to be crucial in the binding of Cav-1. Cav-1 also binds to S630 of the DPP-IV catalytic triad thus regulating the activity of DPP-IV and its inhibitors. <ref name="Puddu">PMID:33935978</ref> |
| - | <scene name='10/1037491/Glp-1r/1'>Y250 and Y252</scene>, were proved to be crucial in the binding of Cav-1. Cav-1 also binds to S630 of the DPP-IV catalytic triad thus regulating the activity of DPP-IV and its inhibitors. <ref name="Puddu">PMID:33935978</ref> | + | |
Current revision
Structure and Function of Dipeptidyl Peptidase IV (DPP-IV) in Humans
| |||||||||||
References
- ↑ 1.0 1.1 Ahrén B. DPP-4 Inhibition and the Path to Clinical Proof. Front Endocrinol (Lausanne). 2019 Jun 19;10:376. PMID:31275243 doi:10.3389/fendo.2019.00376
- ↑ Khalse M, Bhargava A. A Review on Cardiovascular Outcome Studies of Dipeptidyl Peptidase-4 Inhibitors. Indian J Endocrinol Metab. 2018 Sep-Oct;22(5):689-695. PMID:30294582 doi:10.4103/ijem.IJEM_104_18
- ↑ Hocher B, Reichetzeder C, Alter ML. Renal and cardiac effects of DPP4 inhibitors--from preclinical development to clinical research. Kidney Blood Press Res. 2012;36(1):65-84. PMID:22947920 doi:10.1159/000339028
- ↑ Zhong J, Rajagopalan S. Dipeptidyl Peptidase-4 Regulation of SDF-1/CXCR4 Axis: Implications for Cardiovascular Disease. Front Immunol. 2015 Sep 25;6:477. PMID:26441982 doi:10.3389/fimmu.2015.00477
- ↑ Sharma A, Ren X, Zhang H, Pandey GN. Effect of depression and suicidal behavior on neuropeptide Y (NPY) and its receptors in the adult human brain: A postmortem study. Prog Neuropsychopharmacol Biol Psychiatry. 2022 Jan 10;112:110428. PMID:34411658 doi:10.1016/j.pnpbp.2021.110428
- ↑ Ntafam CN, Beutler BD, Harris RD. Incarcerated gravid uterus: A rare but potentially devastating obstetric complication. Radiol Case Rep. 2022 Mar 10;17(5):1583-1586. PMID:35309386 doi:10.1016/j.radcr.2022.02.034
- ↑ Chung KM, Cheng JH, Suen CS, Huang CH, Tsai CH, Huang LH, Chen YR, Wang AH, Jiaang WT, Hwang MJ, Chen X. The dimeric transmembrane domain of prolyl dipeptidase DPP-IV contributes to its quaternary structure and enzymatic activities. Protein Sci. 2010 Sep;19(9):1627-38. PMID:20572019 doi:10.1002/pro.443
- ↑ Abbott CA, McCaughan GW, Levy MT, Church WB, Gorrell MD. Binding to human dipeptidyl peptidase IV by adenosine deaminase and antibodies that inhibit ligand binding involves overlapping, discontinuous sites on a predicted beta propeller domain. Eur J Biochem. 1999 Dec;266(3):798-810. PMID:10583373 doi:10.1046/j.1432-1327.1999.00902.x
- ↑ Dobers J, Grams S, Reutter W, Fan H. Roles of cysteines in rat dipeptidyl peptidase IV/CD26 in processing and proteolytic activity. Eur J Biochem. 2000 Aug;267(16):5093-100. PMID:10931192 doi:10.1046/j.1432-1327.2000.01571.x
- ↑ Metzler WJ, Yanchunas J, Weigelt C, Kish K, Klei HE, Xie D, Zhang Y, Corbett M, Tamura JK, He B, Hamann LG, Kirby MS, Marcinkeviciene J. Involvement of DPP-IV catalytic residues in enzyme-saxagliptin complex formation. Protein Sci. 2008 Feb;17(2):240-50. PMID:18227430 doi:17/2/240
- ↑ 11.0 11.1 Kim BR, Kim HY, Choi I, Kim JB, Jin CH, Han AR. DPP-IV Inhibitory Potentials of Flavonol Glycosides Isolated from the Seeds of Lens culinaris: In Vitro and Molecular Docking Analyses. Molecules. 2018 Aug 10;23(8):1998. PMID:30103438 doi:10.3390/molecules23081998
- ↑ Scott LJ. Sitagliptin: A Review in Type 2 Diabetes. Drugs. 2017 Feb;77(2):209-224. PMID:28078647 doi:10.1007/s40265-016-0686-9
- ↑ Henness S, Keam SJ. Vildagliptin. Drugs. 2006;66(15):1989-2001; discussion 2002-4. PMID:17100408 doi:10.2165/00003495-200666150-00007
- ↑ Garnock-Jones KP. Saxagliptin/Dapagliflozin: A Review in Type 2 Diabetes Mellitus. Drugs. 2017 Mar;77(3):319-330. PMID:28176222 doi:10.1007/s40265-017-0697-1
- ↑ Keating GM. Alogliptin: a review of its use in patients with type 2 diabetes mellitus. Drugs. 2015 May;75(7):777-96. PMID:25855222 doi:10.1007/s40265-015-0385-y
- ↑ Mulvihill EE, Drucker DJ. Pharmacology, physiology, and mechanisms of action of dipeptidyl peptidase-4 inhibitors. Endocr Rev. 2014 Dec;35(6):992-1019. PMID:25216328 doi:10.1210/er.2014-1035
- ↑ Hughes TE, Mone MD, Russell ME, Weldon SC, Villhauer EB. NVP-DPP728 (1-[[[2-[(5-cyanopyridin-2-yl)amino]ethyl]amino]acetyl]-2-cyano-(S)- pyrrolidine), a slow-binding inhibitor of dipeptidyl peptidase IV. Biochemistry. 1999 Sep 7;38(36):11597-603. PMID:10512614 doi:10.1021/bi990852f
- ↑ Banday MZ, Sameer AS, Nissar S. Pathophysiology of diabetes: An overview. Avicenna J Med. 2020 Oct 13;10(4):174-188. PMID:33437689 doi:10.4103/ajm.ajm_53_20
- ↑ Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev. 2007 Oct;87(4):1409-39. PMID:17928588 doi:10.1152/physrev.00034.2006
- ↑ Gilbert MP, Pratley RE. GLP-1 Analogs and DPP-4 Inhibitors in Type 2 Diabetes Therapy: Review of Head-to-Head Clinical Trials. Front Endocrinol (Lausanne). 2020 Apr 3;11:178. PMID:32308645 doi:10.3389/fendo.2020.00178
- ↑ Puddu A, Maggi D. Emerging Role of Caveolin-1 in GLP-1 Action. Front Endocrinol (Lausanne). 2021 Apr 14;12:668012. PMID:33935978 doi:10.3389/fendo.2021.668012
Student Contributors
- Karisma Moll
- Merritt Jugo
- Sam Magnabosco