|
|
| (47 intermediate revisions not shown.) |
| Line 3: |
Line 3: |
| | ---- | | ---- |
| | ==''A. thaliana'' PrRs== | | ==''A. thaliana'' PrRs== |
| - | <StructureSection load='7CS9' size='350' side='right' caption='A. thaliana PrR1 sturcture' scene=''> | + | <StructureSection load='7CSG' size='350' side='right' caption='A. thaliana PrR2 sturcture (PDB code [[7csg]])' scene=''> |
| | | | |
| - | '''1. General Aspects'''
| + | __TOC__ |
| | | | |
| - | Pinoresinol/Lariciresinol Reductases (PLR) are NADPH-dependent reductases that constitute a family of enzymes involved in the lignans biosynthetic pathway. Lignans are a group of nearly 2000 (Xiao et al., 2021) secondary metabolites, formed by dimers of phenylpropanoids, that can be present in all organs of some non-vascular plants, some Tracheophytes, such as Pteridophytes, and Gymnosperms (Markulin et al., 2019). Although their biological role is not yet totally clarified, they are associated mainly with plants defense mechanisms, especially towards herbivores and pathogenic microorganisms (Markulin et al., 2019), providing antibacterial, antifungal and antifeedant effects (Markulin et al., 2019), besides many other impacts.
| + | =='''General Aspects'''== |
| | | | |
| - | Lignans are synthesized by the dimerization of 2 phenylpropanoids: [[Image:Síntese de lignanas menor certo.png]] | + | '''Pinoresinol/Lariciresinol Reductases''' (PLR) or '''pinoresinol reductase''' (PrR) are NADPH-dependent reductases that constitute a family of enzymes involved in the lignans biosynthetic pathway. Lignans are a group of nearly 2000 (Xiao et al., 2021)<ref name="xiao">PMID:33990581</ref> secondary metabolites, formed by dimers of phenylpropanoids, that can be present in all organs of some non-vascular plants, some Tracheophytes, such as Pteridophytes, and Gymnosperms (Markulin et al., 2019)<ref name="markulin">PMID:30895445</ref>. Although their biological role is not yet totally clarified, they are associated mainly with plants defense mechanisms, especially towards herbivores and pathogenic microorganisms (Markulin et al., 2019)<ref name="markulin" />, providing antibacterial, antifungal and antifeedant effects (Markulin et al., 2019)<ref name="markulin" />, besides many other impacts. |
| | | | |
| - | The presence or absence of the 8’,8’ bond between phenylpropanoid monomers differentiate lignans from neolignans, however overall both are associated with many biological activities in humans, due to their structural diversity (Xiao et al., 2021), including antiinflammatory, antioxidant, neuroprotective, antiviral, insecticidal and antitumor proliferation effects (Wang et al., 2022). Given the possibility of positively impacting human health, and their large diversity, lignans are thoroughly explored by the pharmaceutical industry (Markulin et al., 2019). | |
| | | | |
| - | ''Arabidopsis thaliana'' pinoresinol reductase 1 (AtPrR1) and ''A.thaliana'' pinoresinol reductase 2 (AtPrR2) are enzymes from the PLR family. This family of enzymes first reduces pinoresinol to lariciresinol and then lariciresinol to secoisolariciresinol (Nakatsubo et al., 2008). They are very important for many plants and also for biotechnological applications, because their products are key elements for the synthesis of several subclasses of lignans, which make them pivotal enzymes that greatly contribute to the diversity of lignans (Xiao et al., 2021).
| + | Lignans are synthesized by the dimerization of 2 phenylpropanoids: [[Image:Síntese de lignanas menor certo.png]] |
| | | | |
| - | Even so, a study conducted by Nakastsubo and collaborators (2008) showed that the recombinant AtPLRs exhibit a strong preference for pinoresinol, having almost no activity regarding lariciresinol. Because of that difference towards the family of PLRs, AtPLRs have been renamed by these authors as pinoresinol reductases (PrRs). Phylogeny analysis shows Isatis indigotica PLR1 (IiPLR1) holds the biggest relationship to AtPrR, with more than 80% of amino-acid sequence identity. However, IiPLR1 is able to use both pinoresinol and lariciresinol as substrates, meaning that the substrate specificity in AtPrR is due to few aminoacid residues alterations, causing alterations in protein structure enlightened in the next sessions (Xiao et al., 2021).
| + | The presence or absence of the 8’,8’ bond between phenylpropanoid monomers differentiate lignans from neolignans, however overall both are associated with many biological activities in humans, due to their structural diversity (Xiao et al., 2021)<ref name="xiao" />, including antiinflammatory, antioxidant, neuroprotective, antiviral, insecticidal and antitumor proliferation effects (Wang et al., 2022)<ref>PMID:35842031</ref>. Given the possibility of positively impacting human health, and their large diversity, lignans are thoroughly explored by the pharmaceutical industry (Markulin et al., 2019)<ref name="markulin" />. |
| | | | |
| - | ----
| + | ''Arabidopsis thaliana'' pinoresinol reductase 1 (AtPrR1) and ''A.thaliana'' pinoresinol reductase 2 (AtPrR2) are enzymes from the PLR family. This family of enzymes first reduces pinoresinol to lariciresinol and then lariciresinol to secoisolariciresinol (Nakatsubo et al., 2008)<ref>PMID:18347017</ref>. They are very important for many plants and also for biotechnological applications, because their products are key elements for the synthesis of several subclasses of lignans, which make them pivotal enzymes that greatly contribute to the diversity of lignans (Xiao et al., 2021)<ref name="xiao" />. |
| | | | |
| - | '''2. Protein Structure'''
| + | Even so, a study conducted by Nakastsubo and collaborators (2008) showed that the recombinant AtPLRs exhibit a strong preference for pinoresinol, having almost no activity regarding lariciresinol. Because of that difference towards the family of PLRs, AtPLRs have been renamed by these authors as pinoresinol reductases (PrRs). Phylogeny analysis shows Isatis indigotica PLR1 (IiPLR1) holds the biggest relationship to AtPrR, with more than 80% of amino-acid sequence identity. However, IiPLR1 is able to use both pinoresinol and lariciresinol as substrates, meaning that the substrate specificity in AtPrR is due to few aminoacid residues alterations, causing alterations in protein structure enlightened in the next sessions (Xiao et al., 2021)<ref name="xiao" />. |
| | | | |
| - | At the N-terminal end, each subunit of PrR contains a NADPH binding domain (NBD) formed by seven β-strands surrounded by six α-helices. The C-terminal end comprises the substrate binding domains (SBD), which is formed by two β-strands and five α-helices. In NDB, α-helices are larger than in SBD. Between both domains is a large groove containing a positively charged region that associates with NBD and a hydrophobic region that associates with SBD. Overall, the catalysis process consists of the binding of free NADPH to NBD, followed by binding of pinoresinol to SBD. The substrate is reduced and lariciresinol is released (Xiao et al., 2021).
| + | [[Image:Amino-acid sequence alignment of IiPLR and AtPrRs.png]] |
| | | | |
| - | A flexible loop structure is formed by β-strand 4 (β4) and acts as a switch to control the binding of NADPH and release of NADP+ at NBD: without ligand β4 is disordered, but when PrR is binded with NADP+and pinoresinol it forms a well defined loop. Both monomer loops form a twisted “8” shape that covers NBD and SBD. Furthermore, NADPH binding causes the β2 loop to move slightly towards the coenzyme. NADP+ interacts with PrR grooves through hydrogen bonds and hydrophobic interactions with the NADPH-binding domain (GXXGXXG) and neighbor amino-acid residues (Xiao et al. 2021), being Phe166 responsible for the its stabilization (Markulin et al., 2019).
| + | '''IMAGE 1:'''Amino-acid sequence alignment of IiPLR and AtPrRs. The secondary structure elements of IiPLR1 are placed on top. The GXXGXXG motif is indicated with blue solid lines. Regions constituting dimer interfaces of AtPrRs from Mol-A and Mol-B are indicated with green and cyan solid lines, respectively. Residues that may participate in chiral selection are indicated with yellow triangles. Species are: Ii, Isatis indigotica; At, Arabidopsis thaliana. Available in: Xiao et al., 2021<ref name="xiao" />. |
| | + | ---- |
| | | | |
| - | Pinoresinol binds to the hydrophobic portion of PrR grooves, being stabilized by β2 and α-helice 10 (α10). The binding of pinoresinol to one monomer induces a conformational change in the neighboring monomer that helps stabilize the substrate, resulting in a tighter bond. The substrate’s ends form hydrogen bonds with PrP Met125 and Gly178 residues, hence pinoresinol is bound to the enzyme as a straight chain. With this conformation, both molecules' hydrophobic regions are aligned (Xiao et al., 2021). Moreover, pinoresinol binds in such a way that its furan ring is facing the nicotinamide ring of the bound NADPH (Markulin et al., 2019), which allows the transference of an hydrogen atom to the substrate, reducing it to make one molecule of lariciresinol (Xiao et al., 2021). | |
| | | | |
| - | ----
| + | =='''Protein Structure'''== |
| | | | |
| - | '''3. Substrate selectivity''' | + | At the N-terminal end, <scene name='10/1050287/Monomero/1'>each subunit of PrR</scene> (proteins that work as homodimers) contains a <scene name='10/1050287/Nbd/1'>NADPH binding domain (NBD - in blue)</scene> formed by seven β-strands surrounded by six α-helices. The C-terminal end comprises the <scene name='10/1050287/Sbd/1'>substrate binding domain (SBD - in pink)</scene>, which is formed by two β-strands and five α-helices. In NDB, α-helices are larger than in SBD. Between both domains is a large groove containing a positively charged region that associates with NBD and a hydrophobic region that associates with SBD. Overall, the catalysis process consists of the binding of free NADPH to NBD, followed by binding of pinoresinol to SBD. The substrate is reduced and lariciresinol is released (Xiao et al., 2021)<ref name="xiao" />. |
| | | | |
| - | In contrast with AtPrR, IiPLR β4 loops are disorder regardless of the ligand state of the protein, indicating an influence in substrate selectivity and catalysis. The β4 loop amino-acid sequence differs by one residue between IiPLR and AtPrR: at the C-terminal end of the loop, Ser98 is substituted by Asn98 in ''A. thaliana''. The interaction between Asn98 and NADPH results in a position shift of the asparagine side chain, physically limiting the β4 loop's movement in PrR. Furthermore, due to a change in the amino-acid sequence from Val46 to Leu46 in PrR1, SBD is condensed and tighter, limiting the entrance and orientation of the substrate. In PrR2, the same result is observed, and although there is no change in aminoacid residue, Val46 is pushed further into SBD, possibly owing to different dimer orientation. Thus, it is suggested that aminoacid residues at position 46 and 98 are essential for substrate selectivity (Xiao et al., 2021).
| + | A flexible loop structure is formed by β-strand 4 (β4) and acts as a switch to control the binding of NADPH and release of NADP+ at NBD: without ligand β4 is disordered, but when PrR is binded with NADPH and pinoresinol it forms a well defined loop. Both monomer loops form a twisted “8” shape that covers NBD and SBD. Furthermore, NADPH binding causes the β2 loop to move slightly towards the coenzyme. NADPH interacts with PrR grooves through hydrogen bonds and hydrophobic interactions with the NADPH-binding domain (<scene name='10/1050287/Gxxgxxg/4'>GXXGXXG</scene>) and neighbor amino-acid residues (Xiao et al. 2021)<ref name="xiao" />, being <scene name='10/1050287/Phe166/4'>Phe166</scene> responsible for the its stabilization (Markulin et al., 2019)<ref name="markulin" />. |
| | + | |
| | + | Pinoresinol binds to the hydrophobic portion of PrR grooves, being stabilized by β2 and α-helice 10 (α10). The binding of pinoresinol to one monomer induces a conformational change in the neighboring monomer that helps stabilize the substrate, resulting in a tighter bond. <scene name='10/1050287/Met125_e_gly178/1'>The substrate’s ends form hydrogen bonds with PrR Met125 and Gly178 residues</scene>, hence pinoresinol is bound to the enzyme as a straight chain. With this conformation, both molecules' hydrophobic regions are aligned (Xiao et al., 2021)<ref name="xiao" />. Moreover, pinoresinol binds in such a way that its <scene name='10/1050287/Nadph_e_pinoresinol/1'>furan ring is facing the nicotinamide ring</scene> of the bound NADPH (Markulin et al., 2019)<ref name="markulin" />, which allows the transference of an hydrogen atom to the substrate, reducing it to make one molecule of lariciresinol (Xiao et al., 2021)<ref name="xiao" />. |
| | | | |
| | ---- | | ---- |
| | | | |
| - | </StructureSection>
| + | =='''Substrate selectivity'''== |
| | | | |
| - | '''4. AtPrR expression''' | + | In contrast with AtPrR, IiPLR β4 loops are disorder regardless of the ligand state of the protein, indicating an influence in substrate selectivity and catalysis. The β4 loop amino-acid sequence differs by one residue between IiPLR and AtPrR: at the C-terminal end of the loop, Ser98 is substituted by Asn98 in ''A. thaliana''. The interaction between Asn98 and NADPH results in a position shift of the asparagine side chain, physically limiting the β4 loop's movement in PrR. Furthermore, due to a change in the amino-acid sequence from Val46 to Leu46 in PrR1, SBD is condensed and tighter, limiting the entrance and orientation of the substrate. In PrR2, the same result is observed, and although there is no change in aminoacid residue, Val46 is pushed further into SBD, possibly owing to different dimer orientation. Thus, it is suggested that aminoacid residues at position 46 and 98 are essential for substrate selectivity (Xiao et al., 2021)<ref name="xiao" />. |
| | | | |
| - | Understanding the pattern of expression of the PLR family is key in the study of lignans, considering that the diversity of these secondary metabolites is due to the action of these enzymes, but also to their expression pattern (Xiao et al., 2021; Markulin et al., 2019), which dictates where in the plants and when a certain lignan will be produced.
| + | [[Image:425 2019 3137 Fig2 HTML.png|600px]] |
| | + | '''IMAGE 2:'''Enantiospecifcity of characterized pinoresinol–lariciresinol reductases. Available in: Markulin et al., 2019<ref name="markulin" />. |
| | | | |
| - | The pattern of expression of the PLRs varies from plant to plant and it can be altered by different stresses and hormones (Markunil et al., 2019). In the case of Arabidopsis thaliana, both PrR1 and PrR2 are expressed in root, but only PrR1 is also expressed in stem. Zhao and collaborators (2014) have shown by co-expression analysis that PrR1 might be related to the deposition of secondary cell wall, as it clustered with genes related to: lignification, hemicellulose biosynthesis, cellulose synthesis, etc. This possibility was reinforced by transactivation assays in which the trans-activators factors MYB46 and SND1, both related to the deposition of secondary cell wall, interacted with the PrR1 promoter. On other hand, PrR2 clustered with a different set of genes, been close to genes not related to secondary cell wall deposition, as well to genes related to lignification and to the formation of the casparian strip.
| + | =='''AtPrR expression'''== |
| | | | |
| - | What might look contradictory is the fact that PrR1 and PrR2 are coexpressed with lignin related genes and that pinoresinol, the first substrate of the PLR enzymes, is synthetized by the dimerization of two coniferyl alcohols, onde of the monomers that constitute the lignin polymer. We could imagine that the production of lignans would compete for substrates with the biosynthesis of lignin, and therefore the coexpression mentioned would be questionable. Zhao and collaborators (2014) have shown that mutants of A. thaliana without the function of the PrR1 enzyme show lower content of lignin, meaning that, somehow the production of lignans is important for lignification and related to lignin polymerization.
| + | Understanding the pattern of expression of the PLR family is key in the study of lignans, considering that the diversity of these secondary metabolites is due to the action of these enzymes, but also to their expression pattern (Xiao et al., 2021; Markulin et al., 2019)<ref name="xiao" /><ref name="markulin" />, which dictates where in the plants and when a certain lignan will be produced. |
| | | | |
| - | ---- | + | The pattern of expression of the PLRs varies from plant to plant and it can be altered by different stresses and hormones (Markulin et al., 2019)<ref name="markulin" />. In the case of Arabidopsis thaliana, both PrR1 and PrR2 are expressed in root, but only PrR1 is also expressed in stem. Zhao and collaborators (2014)<ref name="zhao">PMID:25107662</ref> have shown by co-expression analysis that PrR1 might be related to the deposition of secondary cell wall, as it clustered with genes related to: lignification, hemicellulose biosynthesis, cellulose synthesis, etc. This possibility was reinforced by transactivation assays in which the trans-activators factors MYB46 and SND1, both related to the deposition of secondary cell wall, interacted with the PrR1 promoter. On other hand, PrR2 clustered with a different set of genes, been close to genes not related to secondary cell wall deposition, as well to genes related to lignification and to the formation of the casparian strip. |
| | | | |
| - | '''5. References'''
| + | What might look contradictory is the fact that PrR1 and PrR2 are coexpressed with lignin related genes and that pinoresinol, the first substrate of the PLR enzymes, is synthetized by the dimerization of two coniferyl alcohols, onde of the monomers that constitute the lignin polymer. We could imagine that the production of lignans would compete for substrates with the biosynthesis of lignin, and therefore the coexpression mentioned would be questionable. Zhao and collaborators (2014)<ref name="zhao" />.have shown that mutants of A. thaliana without the function of the PrR1 enzyme show lower content of lignin, meaning that, somehow the production of lignans is important for lignification and related to lignin polymerization. |
| | | | |
| - | MARKULIN, L. et al. Pinoresinol–lariciresinol reductases, key to the lignan synthesis in plants. Planta, v. 249, n. 6, p. 1695–1714, 20 mar. 2019. PMID: 30895445. DOI: https://doi.org/10.1007/s00425-019-03137-y.
| + | =='''3D structures of Pinoresinol/Lariciresinol Reductase'''== |
| | | | |
| - | TOMOYUKI NAKATSUBO et al. Characterization of Arabidopsis thaliana Pinoresinol Reductase, a New Type of Enzyme Involved in Lignan Biosynthesis. Journal of Biological Chemistry, v. 283, n. 23, p. 15550–15557, 1 jun. 2008. PMID: 18347017. DOI: https://doi.org/10.1074%2Fjbc.M801131200.
| + | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} |
| | | | |
| - | WARD, R. Lignans, neolignans and related compounds. v. 16, n. 1, p. 75–96, 1 jan. 1999.
| + | [[7cs9]] – AtPrR1 -''Arabidopsis thaliana''<br /> |
| | + | [[7csa]] – AtPrR1 + NADP <br /> |
| | + | [[7csd]], [[7cse]] – AtPrR1 + NADP + lariciresinol <br /> |
| | + | [[7csf]] – AtPrR1 + NADP + secoisolariciresinol <br /> |
| | + | [[7csb]], [[7csc]] – AtPrR1 + NADP + pinoresinol <br /> |
| | + | [[7csg]] – AtPrR2 <br /> |
| | + | [[7csh]] – ItPrR2 + NADP + pinoresinol <br /> |
| | + | [[1qyd]] – PrR - giant arborvitae<br /> |
| | + | [[7cs2]] – ItPrR1 -''Isatis tinctoria''<br /> |
| | + | [[7cs3]] – ItPrR1 + NADP <br /> |
| | + | [[7cs6]] – ItPrR1 + NADP + lariciresinol <br /> |
| | + | [[7cs4]], [[7cs5]] – ItPrR1 + NADP + pinoresinol <br /> |
| | + | [[7cs7]], [[7cs8]] – ItPrR1 + NADP + secoisolariciresinol <br /> |
| | | | |
| - | XIAO, Y. et al. Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases. Nature communications, v. 12, n. 1, 14 maio 2021. PMID: 33990581. DOI: https://doi.org/10.1038/s41467-021-23095-y.
| + | =='''References'''== |
| | + | <references /> |
| | | | |
| - | ZHAO, Q. et al. Pinoresinol reductase 1 impacts lignin distribution during secondary cell wall biosynthesis in Arabidopsis. Phytochemistry, v. 112, p. 170–178, 1 abr. 2015. PMID: 25107662. DOI: https://doi.org/10.1016/j.phytochem.2014.07.008.
| + | WARD, R. Lignans, neolignans and related compounds. v. 16, n. 1, p. 75–96, 1 jan. 1999. doi: https://doi.org/10.1039/A705992B |
| | + | |
| | + | </StructureSection> |
| | + | [[Category:Topic Page]] |
Página sobre a PrR1 e PrR2 de Arabidopsis feita por alunos da Biologia da USP São Paulo.
|
General Aspects
Pinoresinol/Lariciresinol Reductases (PLR) or pinoresinol reductase (PrR) are NADPH-dependent reductases that constitute a family of enzymes involved in the lignans biosynthetic pathway. Lignans are a group of nearly 2000 (Xiao et al., 2021)[1] secondary metabolites, formed by dimers of phenylpropanoids, that can be present in all organs of some non-vascular plants, some Tracheophytes, such as Pteridophytes, and Gymnosperms (Markulin et al., 2019)[2]. Although their biological role is not yet totally clarified, they are associated mainly with plants defense mechanisms, especially towards herbivores and pathogenic microorganisms (Markulin et al., 2019)[2], providing antibacterial, antifungal and antifeedant effects (Markulin et al., 2019)[2], besides many other impacts.
Lignans are synthesized by the dimerization of 2 phenylpropanoids: 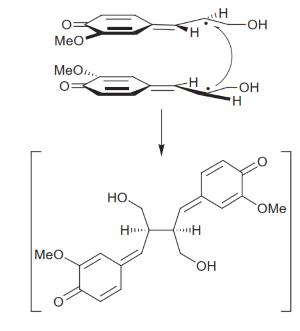
The presence or absence of the 8’,8’ bond between phenylpropanoid monomers differentiate lignans from neolignans, however overall both are associated with many biological activities in humans, due to their structural diversity (Xiao et al., 2021)[1], including antiinflammatory, antioxidant, neuroprotective, antiviral, insecticidal and antitumor proliferation effects (Wang et al., 2022)[3]. Given the possibility of positively impacting human health, and their large diversity, lignans are thoroughly explored by the pharmaceutical industry (Markulin et al., 2019)[2].
Arabidopsis thaliana pinoresinol reductase 1 (AtPrR1) and A.thaliana pinoresinol reductase 2 (AtPrR2) are enzymes from the PLR family. This family of enzymes first reduces pinoresinol to lariciresinol and then lariciresinol to secoisolariciresinol (Nakatsubo et al., 2008)[4]. They are very important for many plants and also for biotechnological applications, because their products are key elements for the synthesis of several subclasses of lignans, which make them pivotal enzymes that greatly contribute to the diversity of lignans (Xiao et al., 2021)[1].
Even so, a study conducted by Nakastsubo and collaborators (2008) showed that the recombinant AtPLRs exhibit a strong preference for pinoresinol, having almost no activity regarding lariciresinol. Because of that difference towards the family of PLRs, AtPLRs have been renamed by these authors as pinoresinol reductases (PrRs). Phylogeny analysis shows Isatis indigotica PLR1 (IiPLR1) holds the biggest relationship to AtPrR, with more than 80% of amino-acid sequence identity. However, IiPLR1 is able to use both pinoresinol and lariciresinol as substrates, meaning that the substrate specificity in AtPrR is due to few aminoacid residues alterations, causing alterations in protein structure enlightened in the next sessions (Xiao et al., 2021)[1].
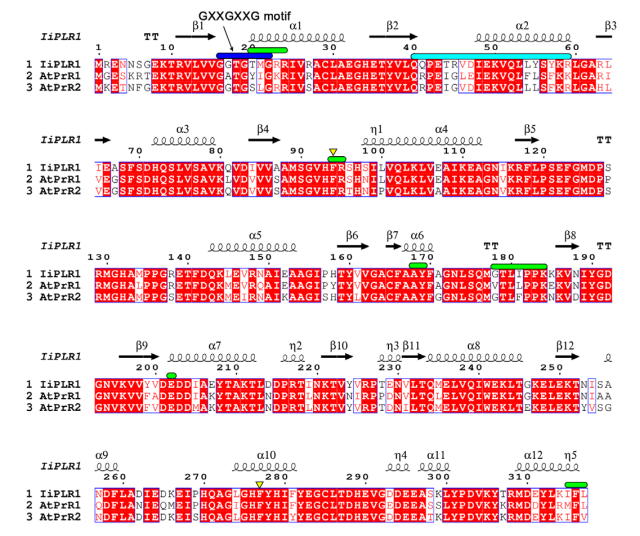
IMAGE 1:Amino-acid sequence alignment of IiPLR and AtPrRs. The secondary structure elements of IiPLR1 are placed on top. The GXXGXXG motif is indicated with blue solid lines. Regions constituting dimer interfaces of AtPrRs from Mol-A and Mol-B are indicated with green and cyan solid lines, respectively. Residues that may participate in chiral selection are indicated with yellow triangles. Species are: Ii, Isatis indigotica; At, Arabidopsis thaliana. Available in: Xiao et al., 2021[1].
Protein Structure
At the N-terminal end, (proteins that work as homodimers) contains a formed by seven β-strands surrounded by six α-helices. The C-terminal end comprises the , which is formed by two β-strands and five α-helices. In NDB, α-helices are larger than in SBD. Between both domains is a large groove containing a positively charged region that associates with NBD and a hydrophobic region that associates with SBD. Overall, the catalysis process consists of the binding of free NADPH to NBD, followed by binding of pinoresinol to SBD. The substrate is reduced and lariciresinol is released (Xiao et al., 2021)[1].
A flexible loop structure is formed by β-strand 4 (β4) and acts as a switch to control the binding of NADPH and release of NADP+ at NBD: without ligand β4 is disordered, but when PrR is binded with NADPH and pinoresinol it forms a well defined loop. Both monomer loops form a twisted “8” shape that covers NBD and SBD. Furthermore, NADPH binding causes the β2 loop to move slightly towards the coenzyme. NADPH interacts with PrR grooves through hydrogen bonds and hydrophobic interactions with the NADPH-binding domain () and neighbor amino-acid residues (Xiao et al. 2021)[1], being responsible for the its stabilization (Markulin et al., 2019)[2].
Pinoresinol binds to the hydrophobic portion of PrR grooves, being stabilized by β2 and α-helice 10 (α10). The binding of pinoresinol to one monomer induces a conformational change in the neighboring monomer that helps stabilize the substrate, resulting in a tighter bond. , hence pinoresinol is bound to the enzyme as a straight chain. With this conformation, both molecules' hydrophobic regions are aligned (Xiao et al., 2021)[1]. Moreover, pinoresinol binds in such a way that its of the bound NADPH (Markulin et al., 2019)[2], which allows the transference of an hydrogen atom to the substrate, reducing it to make one molecule of lariciresinol (Xiao et al., 2021)[1].
Substrate selectivity
In contrast with AtPrR, IiPLR β4 loops are disorder regardless of the ligand state of the protein, indicating an influence in substrate selectivity and catalysis. The β4 loop amino-acid sequence differs by one residue between IiPLR and AtPrR: at the C-terminal end of the loop, Ser98 is substituted by Asn98 in A. thaliana. The interaction between Asn98 and NADPH results in a position shift of the asparagine side chain, physically limiting the β4 loop's movement in PrR. Furthermore, due to a change in the amino-acid sequence from Val46 to Leu46 in PrR1, SBD is condensed and tighter, limiting the entrance and orientation of the substrate. In PrR2, the same result is observed, and although there is no change in aminoacid residue, Val46 is pushed further into SBD, possibly owing to different dimer orientation. Thus, it is suggested that aminoacid residues at position 46 and 98 are essential for substrate selectivity (Xiao et al., 2021)[1].
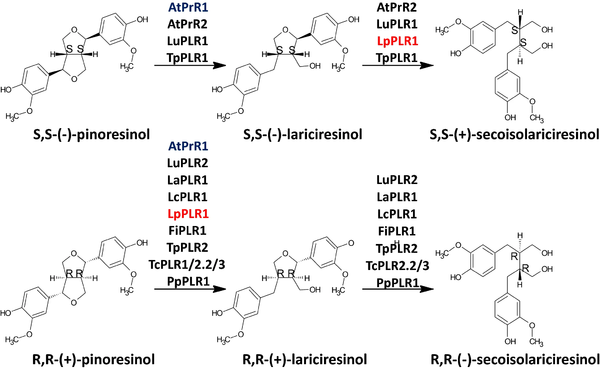
IMAGE 2:Enantiospecifcity of characterized pinoresinol–lariciresinol reductases. Available in: Markulin et al., 2019[2].
AtPrR expression
Understanding the pattern of expression of the PLR family is key in the study of lignans, considering that the diversity of these secondary metabolites is due to the action of these enzymes, but also to their expression pattern (Xiao et al., 2021; Markulin et al., 2019)[1][2], which dictates where in the plants and when a certain lignan will be produced.
The pattern of expression of the PLRs varies from plant to plant and it can be altered by different stresses and hormones (Markulin et al., 2019)[2]. In the case of Arabidopsis thaliana, both PrR1 and PrR2 are expressed in root, but only PrR1 is also expressed in stem. Zhao and collaborators (2014)[5] have shown by co-expression analysis that PrR1 might be related to the deposition of secondary cell wall, as it clustered with genes related to: lignification, hemicellulose biosynthesis, cellulose synthesis, etc. This possibility was reinforced by transactivation assays in which the trans-activators factors MYB46 and SND1, both related to the deposition of secondary cell wall, interacted with the PrR1 promoter. On other hand, PrR2 clustered with a different set of genes, been close to genes not related to secondary cell wall deposition, as well to genes related to lignification and to the formation of the casparian strip.
What might look contradictory is the fact that PrR1 and PrR2 are coexpressed with lignin related genes and that pinoresinol, the first substrate of the PLR enzymes, is synthetized by the dimerization of two coniferyl alcohols, onde of the monomers that constitute the lignin polymer. We could imagine that the production of lignans would compete for substrates with the biosynthesis of lignin, and therefore the coexpression mentioned would be questionable. Zhao and collaborators (2014)[5].have shown that mutants of A. thaliana without the function of the PrR1 enzyme show lower content of lignin, meaning that, somehow the production of lignans is important for lignification and related to lignin polymerization.
3D structures of Pinoresinol/Lariciresinol Reductase
Updated on 02-January-2025
7cs9 – AtPrR1 -Arabidopsis thaliana
7csa – AtPrR1 + NADP
7csd, 7cse – AtPrR1 + NADP + lariciresinol
7csf – AtPrR1 + NADP + secoisolariciresinol
7csb, 7csc – AtPrR1 + NADP + pinoresinol
7csg – AtPrR2
7csh – ItPrR2 + NADP + pinoresinol
1qyd – PrR - giant arborvitae
7cs2 – ItPrR1 -Isatis tinctoria
7cs3 – ItPrR1 + NADP
7cs6 – ItPrR1 + NADP + lariciresinol
7cs4, 7cs5 – ItPrR1 + NADP + pinoresinol
7cs7, 7cs8 – ItPrR1 + NADP + secoisolariciresinol
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Xiao Y, Shao K, Zhou J, Wang L, Ma X, Wu D, Yang Y, Chen J, Feng J, Qiu S, Lv Z, Zhang L, Zhang P, Chen W. Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases. Nat Commun. 2021 May 14;12(1):2828. PMID:33990581 doi:10.1038/s41467-021-23095-y
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Markulin L, Corbin C, Renouard S, Drouet S, Gutierrez L, Mateljak I, Auguin D, Hano C, Fuss E, Lainé E. Pinoresinol-lariciresinol reductases, key to the lignan synthesis in plants. Planta. 2019 Jun;249(6):1695-1714. PMID:30895445 doi:10.1007/s00425-019-03137-y
- ↑ Wang LX, Wang HL, Huang J, Chu TZ, Peng C, Zhang H, Chen HL, Xiong YA, Tan YZ. Review of lignans from 2019 to 2021: Newly reported compounds, diverse activities, structure-activity relationships and clinical applications. Phytochemistry. 2022 Oct;202:113326. PMID:35842031 doi:10.1016/j.phytochem.2022.113326
- ↑ Nakatsubo T, Mizutani M, Suzuki S, Hattori T, Umezawa T. Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J Biol Chem. 2008 Jun 6;283(23):15550-7. PMID:18347017 doi:10.1074/jbc.M801131200
- ↑ 5.0 5.1 Zhao Q, Zeng Y, Yin Y, Pu Y, Jackson LA, Engle NL, Martin MZ, Tschaplinski TJ, Ding SY, Ragauskas AJ, Dixon RA. Pinoresinol reductase 1 impacts lignin distribution during secondary cell wall biosynthesis in Arabidopsis. Phytochemistry. 2015 Apr;112:170-8. PMID:25107662 doi:10.1016/j.phytochem.2014.07.008
WARD, R. Lignans, neolignans and related compounds. v. 16, n. 1, p. 75–96, 1 jan. 1999. doi: https://doi.org/10.1039/A705992B
|



