Journal:Acta Cryst D:S2059798324005229
From Proteopedia
(Difference between revisions)

| (29 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <StructureSection load='' size='450' side='right' scene=' | + | <StructureSection load='' size='450' side='right' scene='10/1050322/Cv/1' caption=''> |
===Factors affecting macromolecule orientations in thin films formed in cryo-EM=== | ===Factors affecting macromolecule orientations in thin films formed in cryo-EM=== | ||
<big>Swati Yadav and Vinothkumar Kutti Ragunath</big> <ref>doi: 10.1107/S2059798324005229</ref> | <big>Swati Yadav and Vinothkumar Kutti Ragunath</big> <ref>doi: 10.1107/S2059798324005229</ref> | ||
| Line 5: | Line 5: | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
Single particle cryo-EM has become a routine and indispensable technique in structural biology, where macromolecules are imaged with electrons as single particles and subsequently averaged to obtain a high-resolution 3D reconstruction. Single particle cryo-EM reconstruction utilizes the projection slice theorem to build a 3D map of a 3D object using 2D projections, generated when electrons transmitted through a thin specimen are recorded by a detector. The resolution and quality of the final map depend on many factors including heterogeneity in the specimen, adequate sampling of views and signal-to-noise of the images. | Single particle cryo-EM has become a routine and indispensable technique in structural biology, where macromolecules are imaged with electrons as single particles and subsequently averaged to obtain a high-resolution 3D reconstruction. Single particle cryo-EM reconstruction utilizes the projection slice theorem to build a 3D map of a 3D object using 2D projections, generated when electrons transmitted through a thin specimen are recorded by a detector. The resolution and quality of the final map depend on many factors including heterogeneity in the specimen, adequate sampling of views and signal-to-noise of the images. | ||
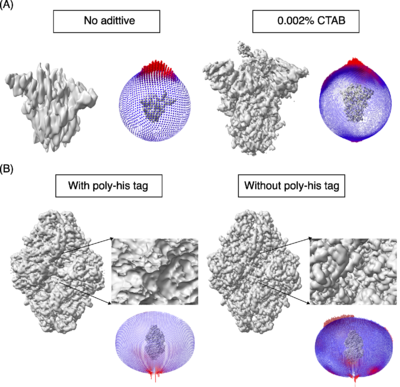
| - | A key step in this process is to obtain a thin film of macromolecules (purified or as a mixture) in ice and the most routinely used method for obtaining such thin films was developed by Dubochet and his colleagues (Dubochet et al., 1988). Obtaining the optimal conditions for freezing of a given macromolecule requires testing several parameters. At the moment, it is one of the rate-limiting steps in obtaining high-resolution maps by cryo-EM. During the freezing process, the macromolecules can encounter and interact with the air-water interface, sometimes resulting in a tendency to adapt one or more preferential orientations along with denaturation and dissociation in the case of multi-subunit complexes (D’Imprima et al., 2019; Noble et al., 2018; Glaeser and Han, 2017). The 3D maps reconstructed from preferentially oriented macromolecules exhibit stretching of densities in one direction (also called anisotropy, where with enough particles the resolution will be high but the maps are not easily interpretable). One such example is the spike protein shown in Figure 1A, which illustrates the effects of orientation bias on the final reconstruction. | + | A key step in this process is to obtain a thin film of macromolecules (purified or as a mixture) in ice and the most routinely used method for obtaining such thin films was developed by Dubochet and his colleagues (Dubochet ''et al.,'' 1988<ref name="Dubochet1">PMID:3043536</ref>). Obtaining the optimal conditions for freezing of a given macromolecule requires testing several parameters. At the moment, it is one of the rate-limiting steps in obtaining high-resolution maps by cryo-EM. During the freezing process, the macromolecules can encounter and interact with the air-water interface, sometimes resulting in a tendency to adapt one or more preferential orientations along with denaturation and dissociation in the case of multi-subunit complexes (D’Imprima ''et al.,'' 2019<ref name="D’Imprima">PMID:30932812</ref>; Noble ''et al.,'' 2018<ref name="Noble">PMID: 30250056</ref>; Glaeser and Han, 2017<ref name="Glaeser">PMID: 28781996</ref>). The 3D maps reconstructed from preferentially oriented macromolecules exhibit stretching of densities in one direction (also called anisotropy, where with enough particles the resolution will be high but the maps are not easily interpretable). One such example is the spike protein shown in Figure 1A, which illustrates the effects of orientation bias on the final reconstruction. |
| - | There are multiple ways to address orientation bias including the use of support layers like carbon or graphene, tilting of the stage and more commonly, the use of surfactants or detergents as small molecule additives during grid preparation (Liu and Wang, 2023). There are a number of macromolecules where surfactants have been used to overcome the preferred orientation problem. In this work, we asked if an informed decision regarding the grid freezing conditions can be made based on the properties of the macromolecule. | + | [[Image:Figure1RR5238.png|left|thumb|400px]] |
| - | We studied macromolecules of different sizes and symmetries (125-440 kDa, C1 to D3) and tested a few commonly used surfactants to overcome the orientation bias and observed that many of these surfactants are beneficial. One such example is cationic surfactant CTAB, which causes changes in the orientation distribution of SARS-CoV-2 spike protein as shown in Figure1A. Further, we tested the effect of poly-histidine tag on orientation distribution of macromolecules. Figure 1B shows the impact of the N-terminal poly-his tag on orientation in the case of the E. coli | + | {{Clear}} |
| + | '''Figure 1:''' The models used as reference are PDBs [[8h3d]] and [[6cvm]] for the spike protein and b-galactosidase respectively. A) Spike protein, SARS-COV-2. <scene name='10/1050322/Spike_no_additive/1'>Spike, no additive</scene> ([[8h3d]]). <scene name='10/1050322/Spike_with_ctab/1'>Spike with CTAB</scene> ([[8wzi]]). B) β-galactosidase. | ||
| + | |||
| + | There are multiple ways to address orientation bias including the use of support layers like carbon or graphene, tilting of the stage and more commonly, the use of surfactants or detergents as small molecule additives during grid preparation (Liu and Wang, 2023<ref name="Liu">PMID: 36563741</ref>). There are a number of macromolecules where surfactants have been used to overcome the preferred orientation problem. In this work, we asked if an informed decision regarding the grid freezing conditions can be made based on the properties of the macromolecule. | ||
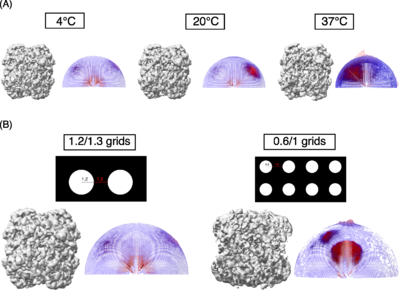
| + | We studied macromolecules of different sizes and symmetries (125-440 kDa, C1 to D3) and tested a few commonly used surfactants to overcome the orientation bias and observed that many of these surfactants are beneficial. One such example is cationic surfactant CTAB, which causes changes in the orientation distribution of SARS-CoV-2 spike protein as shown in Figure1A. Further, we tested the effect of poly-histidine tag on orientation distribution of macromolecules. Figure 1B shows the impact of the N-terminal poly-his tag on orientation in the case of the E. coli β-galactosidase enzyme. We assessed the improvement in orientation distribution with different measures including the orientation distribution plot after 3D refinement (Scheres 2012<ref name="Scheres">PMID: 23000701</ref>), Efficiency of orientation distribution (Naydenova & Russo, 2017<ref name="Naydenova">PMID: 28931821</ref>), 3D-FSC (Tan ''et al.,'' 2018<ref name="Tan">PMID: 28671674</ref>) and the map vs model FSC curve. We also tested the effect of grid hole size and temperature during grid preparation on orientation bias demonstrated using human erythrocyte catalase as an example (Figure 2). | ||
| + | |||
| + | [[Image:Figure2RR5238.png|left|thumb|400px]] | ||
| + | {{Clear}} | ||
| + | '''Figure 2:''' Human erythrocyte catalase. <scene name='10/1050322/Catalase/1'>Catalase at 20 °C</scene> (PDB ID [[8wzj]]). | ||
| + | |||
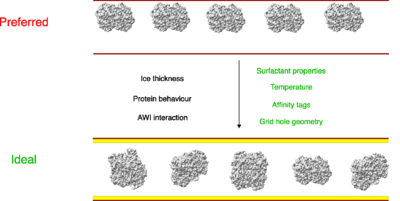
| + | In summary, physical and chemical factors affecting macromolecule behaviour on grids have been studied to overcome the preferred orientation problem (Figure 3). These findings lay a platform in achieving optimal freezing conditions for any given macromolecule in the future. | ||
| + | |||
| + | [[Image:Figure3RR5238.png|left|thumb|400px]] | ||
| + | {{Clear}} | ||
| + | '''Figure 3''' Physical and chemical factors affecting macromolecule behaviour on grids have been studied to overcome the preferred orientation problem. | ||
| + | |||
| + | <scene name='10/1050322/Crppentamer/3'>CRP Pentamer with CTAB</scene> (PDB ID [[8wv4]]). | ||
| + | |||
| + | <scene name='10/1050322/Crpdecamer/1'>CRP decamer with CTAB</scene> (PDB ID [[8wv5]]). | ||
| + | |||
| + | <scene name='10/1050322/Paaz/1'>PaaZ with CTAB at 4 °C</scene> (PDB ID [[8wv6]]). | ||
<b>References</b><br> | <b>References</b><br> | ||
Current revision
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.