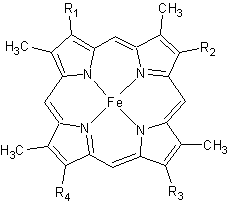
Cytochrome c
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 14: | Line 14: | ||
*'''Cytochrome c557''' and '''cytochrome c558''' are mitochondrial and contain atypical harm-binding sitei<ref> PMID 242319</ref>. | *'''Cytochrome c557''' and '''cytochrome c558''' are mitochondrial and contain atypical harm-binding sitei<ref> PMID 242319</ref>. | ||
*'''Cytochrome cL''' is found in methylotrops. It receives an electron from PQQ cofactor of methanol dehydrogenase to produce formaldehyde<ref> PMID 32627749</ref>. | *'''Cytochrome cL''' is found in methylotrops. It receives an electron from PQQ cofactor of methanol dehydrogenase to produce formaldehyde<ref> PMID 32627749</ref>. | ||
| + | *'''Cytochrome cd1''' icatalyzes the reduction of NO2 to NO and water. | ||
| + | *'''Cytochrome cH''' is the electron donor to the oxidase in methylotrophs<ref> PMID 10386873</ref>. | ||
| + | *'''Cytochrome c XoxG''' is a cytochrome from the lanthanide-dependent methanol dehydrogenase system of methylothrophic bacteria<ref> PMID 31017712</ref>. | ||
See also [[Cytochrome C -Adis]], [[Hemeproteins]], [[Cytochrome C (Hebrew)]], [[Cytochrome C (arabic)]]. | See also [[Cytochrome C -Adis]], [[Hemeproteins]], [[Cytochrome C (Hebrew)]], [[Cytochrome C (arabic)]]. | ||
Current revision
| |||||||||||
References
- ↑ Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001 Nov 2;313(4):903-19. PMID:11697912 doi:10.1006/jmbi.2001.5080
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 Stelter M, Melo AM, Pereira MM, Gomes CM, Hreggvidsson GO, Hjorleifsdottir S, Saraiva LM, Teixeira M, Archer M. A Novel Type of Monoheme Cytochrome c: Biochemical and Structural Characterization at 1.23 A Resolution of Rhodothermus marinus Cytochrome c. Biochemistry. 2008 Oct 15. PMID:18855424 doi:10.1021/bi800999g
- ↑ Axelrod HL, Okamura MY. The structure and function of the cytochrome c2: reaction center electron transfer complex from Rhodobacter sphaeroides. Photosynth Res. 2005;85(1):101-14. PMID:15977062 doi:10.1007/s11120-005-1368-8
- ↑ ElAntak L, Morelli X, Bornet O, Hatchikian C, Czjzek M, Dolla A, Guerlesquin F. The cytochrome c3-[Fe]-hydrogenase electron-transfer complex: structural model by NMR restrained docking. FEBS Lett. 2003 Jul 31;548(1-3):1-4. PMID:12885397
- ↑ Kadziola A, Larsen S. Crystal structure of the dihaem cytochrome c4 from Pseudomonas stutzeri determined at 2.2A resolution. Structure. 1997 Feb 15;5(2):203-16. PMID:9032080
- ↑ Pokkuluri PR, Londer YY, Duke NE, Erickson J, Pessanha M, Salgueiro CA, Schiffer M. Structure of a novel c7-type three-heme cytochrome domain from a multidomain cytochrome c polymer. Protein Sci. 2004 Jun;13(6):1684-92. Epub 2004 May 7. PMID:15133162 doi:10.1110/ps.04626204
- ↑ Roncel M, Kirilovsky D, Guerrero F, Serrano A, Ortega JM. Photosynthetic cytochrome c550. Biochim Biophys Acta. 2012 Aug;1817(8):1152-63. PMID:22289879 doi:10.1016/j.bbabio.2012.01.008
- ↑ Kashey TS, Cowgill JB, McConnell MD, Flores M, Redding KE. Expression and characterization of cytochrome c553 from Heliobacterium modesticaldum. Photosynth Res. 2014 Jun;120(3):291-9. PMID:24557489 doi:10.1007/s11120-014-9982-y
- ↑ Iverson TM, Arciero DM, Hooper AB, Rees DC. High-resolution structures of the oxidized and reduced states of cytochrome c554 from Nitrosomonas europaea. J Biol Inorg Chem. 2001 Apr;6(4):390-7. PMID:11372197
- ↑ Korszun ZR, Salemme FR. Structure of cytochrome c555 of Chlorobium thiosulfatophilum: primitive low-potential cytochrome c. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5244-7. PMID:202947 doi:10.1073/pnas.74.12.5244
- ↑ Pettigrew GW, Aviram I, Schejter A. Physicochemical properties of two atypical cytochromes c, Crithidia cytochrome c-557 and Euglena cytochrome c-558. Biochem J. 1975 Jul;149(1):155-67. PMID:242319 doi:10.1042/bj1490155
- ↑ Ghosh S, Dhanasingh I, Ryu J, Kim SW, Lee SH. Crystal structure of Cytochrome cL from the aquatic methylotrophic bacterium Methylophaga aminisulfidivorans MP(T). J Microbiol Biotechnol. 2020 Jul 3. pii: jmb.2006.06029. doi:, 10.4014/jmb.2006.06029. PMID:32627749 doi:http://dx.doi.org/10.4014/jmb.2006.06029
- ↑ Read J, Gill R, Dales SL, Cooper JB, Wood SP, Anthony C. The molecular structure of an unusual cytochrome c2 determined at 2.0 A; the cytochrome cH from Methylobacterium extorquens. Protein Sci. 1999 Jun;8(6):1232-40. PMID:10386873
- ↑ Featherston ER, Rose HR, McBride MJ, Taylor E, Boal AK, Cotruvo J Jr. Biochemical and structural characterization of XoxG and XoxJ and their roles in activity of the lanthanide-dependent methanol dehydrogenase, XoxF. Chembiochem. 2019 Apr 24. doi: 10.1002/cbic.201900184. PMID:31017712 doi:http://dx.doi.org/10.1002/cbic.201900184
- ↑ 15.0 15.1 15.2 Reedy CJ, Gibney BR. Heme protein assemblies. Chem Rev. 2004 Feb;104(2):617-49. PMID:14871137 doi:10.1021/cr0206115
- ↑ 16.0 16.1 16.2 Ambler RP. Sequence variability in bacterial cytochromes c. Biochim Biophys Acta. 1991 May 23;1058(1):42-7. PMID:1646017
- ↑ Cookson DJ, Moore GR, Pitt RC, Williams RJP, Campbell ID, Ambler RP, Bruschi M, Le Gall J. Structural homology of cytochromes c. Eur J Biochem. 1978 Feb;83(1):261-75.
- ↑ Than ME, Hof P, Huber R, Bourenkov GP, Bartunik HD, Buse G, Soulimane T. Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing. J Mol Biol. 1997 Aug 29;271(4):629-44. PMID:9281430 doi:10.1006/jmbi.1997.1181
- ↑ Soares CM, Baptista AM, Pereira MM, Teixeira M. Investigation of protonatable residues in Rhodothermus marinus caa3 haem-copper oxygen reductase: comparison with Paracoccus denitrificans aa3 haem-copper oxygen reductase. J Biol Inorg Chem. 2004 Mar;9(2):124-34. Epub 2003 Dec 23. PMID:14691678 doi:10.1007/s00775-003-0509-9
- ↑ Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim Biophys Acta. 2001 Jun 1;1505(2-3):185-208. PMID:11334784
- ↑ 21.0 21.1 21.2 21.3 21.4 21.5 Karp, Gerald (2008). Cell and Molecular Biology (5th edition). Hoboken, NJ: John Wiley & Sons. ISBN 978-0470042175.
- ↑ Rajagopal BS, Wilson MT, Bendall DS, Howe CJ, Worrall JA. Structural and kinetic studies of imidazole binding to two members of the cytochrome c (6) family reveal an important role for a conserved heme pocket residue. J Biol Inorg Chem. 2011 Jan 26. PMID:21267610 doi:10.1007/s00775-011-0758-y
- ↑ Morelli X, Czjzek M, Hatchikian CE, Bornet O, Fontecilla-Camps JC, Palma NP, Moura JJ, Guerlesquin F. Structural model of the Fe-hydrogenase/cytochrome c553 complex combining transverse relaxation-optimized spectroscopy experiments and soft docking calculations. J Biol Chem. 2000 Jul 28;275(30):23204-10. PMID:10748163 doi:10.1074/jbc.M909835199
- ↑ Manole A, Kekilli D, Svistunenko DA, Wilson MT, Dobbin PS, Hough MA. Conformational control of the binding of diatomic gases to cytochrome c'. J Biol Inorg Chem. 2015 Mar 20. PMID:25792378 doi:http://dx.doi.org/10.1007/s00775-015-1253-7
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, Melissa Morrison, Adis Hasic