User:Elizabeth Yowell/ SandboxFinal
From Proteopedia
| (7 intermediate revisions not shown.) | |||
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
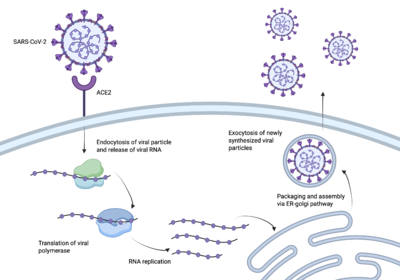
| - | [[Image:Illustration.png|400 px|right|thumb|The process of the SARS-CoV-2 spike protein entering human cells | + | [[Image:Illustration.png|400 px|right|thumb|The process of the SARS-CoV-2 spike protein entering human cells.]] |
SARS-CoV-2 better known as Covid-19 sent the world into a global pandemic due to its rapid transmission and infection <ref name=”WHO”>https://www.who.int/europe/emergencies/situations/covid-19</ref>. In order to create a vaccine, it was essential to understand how SARS-CoV-2 infected us. | SARS-CoV-2 better known as Covid-19 sent the world into a global pandemic due to its rapid transmission and infection <ref name=”WHO”>https://www.who.int/europe/emergencies/situations/covid-19</ref>. In order to create a vaccine, it was essential to understand how SARS-CoV-2 infected us. | ||
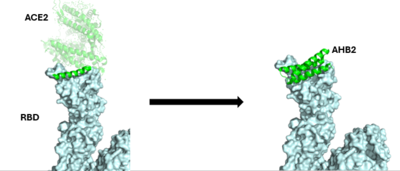
| - | + | The enzyme angiotensin converting enzyme 2 <scene name='10/1077473/Ace2_only/1'>(ACE2)</scene> is attached to our cell membranes and then can be bound to a receptor binding domain <scene name='10/1077473/Rbd_only/2'>(RBD)</scene> <ref name="Borkotoky">DOI:10.1007/s11033-022-08193-4</ref>. The spike protein of SARS-CoV-2 enters our bodies and <scene name='10/1076049/Ace2andrbd/1'>binds to the RBD</scene>, allowing the spike protein access to ACE2 and our host cells <ref name=”Jackson”>DOI:10.1038/s41580-021-00418-x</ref>. Once the spike protein has access to our host cells, it is able to further infect our cells and spread the virus throughout our bodies, causing us to get sick. In order to create an effective vaccine, the pathway between the spike protein and the RBD needed to be [https://www.sciencedirect.com/science/article/pii/S0166354223000190 interrupted]. | |
| - | The enzyme angiotensin converting enzyme 2 <scene name='10/ | + | |
| Line 20: | Line 19: | ||
==Vaccines== | ==Vaccines== | ||
| - | A common way to create vaccines is through the usage of antibodies.<scene name='10/1077473/Antibody_overview/2'>Antibodies</scene> are inserted into the body that mimic ACE2, due to the similarities with ACE2, when COVID-19 spike protein enters the body, they <scene name='10/1077473/Antibody/ | + | A common way to create vaccines is through the usage of antibodies.<scene name='10/1077473/Antibody_overview/2'>Antibodies</scene> are inserted into the body that mimic ACE2, due to the similarities with ACE2, when COVID-19 spike protein enters the body, they <scene name='10/1077473/Antibody/4'>bind</scene> to these ACE2 mimicking antibodies that create a 90% neutralizing response for targeting the RBD. With this being said, this method of treatment is difficult for long term use due to the evolution of the viral cells <ref name="Zhang">DOI:10.1016/S2666-5247(23)00011-3</ref>. |
| - | == | + | ==Inhibitor Development== |
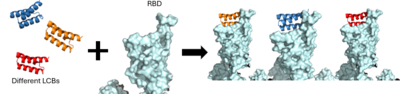
Protein inhibitors were thought of as a new idea for creating vaccines due to their smaller size and better stability compared to antibody vaccines<ref name="Cao">DOI:10.1126/science.abd9909</ref>. These protein inhibitors are also referred to as mini-binders, they interact with the ACE2 receptor binding domain, <scene name='10/1075220/Spikeblockedbyminibinder/2'>preventing association of the viral cell with ACE-2.</scene> | Protein inhibitors were thought of as a new idea for creating vaccines due to their smaller size and better stability compared to antibody vaccines<ref name="Cao">DOI:10.1126/science.abd9909</ref>. These protein inhibitors are also referred to as mini-binders, they interact with the ACE2 receptor binding domain, <scene name='10/1075220/Spikeblockedbyminibinder/2'>preventing association of the viral cell with ACE-2.</scene> | ||
| Line 50: | Line 49: | ||
==Binding Site and Interactions== | ==Binding Site and Interactions== | ||
| - | + | <scene name='10/1075219/Lcb1/1'>LCB1 binding site</scene> | |
| - | + | <scene name='10/1075219/Lcb3/2'>LCB3 binding site</scene> | |
| - | + | <scene name='10/1075219/Ahb2/1'>AHB2 binding site</scene> | |
| - | + | <scene name='10/1075219/Ace2/1'>ACE2 binding site</scene> | |
| - | + | ==Inhibitor Differences== | |
| - | [[Image:Comparison.jpeg|400 px|left|thumb|The sequence differences between ACE2, AHB2, | + | [[Image:Comparison.jpeg|400 px|left|thumb|The sequence differences between ACE2, AHB2, LCB1and LCB3.]] |
Current revision
Contents |
Engineered Protein Inhibitors for SARS-CoV-2 Entry
| |||||||||||
References
Cao, L., Goreshnik, I., Coventry, B., Case, J.B., Miller, L., Kozodoy, L., Chen, R.E., Carter, L., Walls, A.C., Park, Y., Strauch, E., Stewart, L., Diamond, M.S., Veesler, D., & Baker, D. De novo design of picomolar SARS-CoV-2 mini protein inhibitors. Science 370, 426-431 (2020). https://doi.org/10.1126/science.abd9909
https://www.who.int/europe/emergencies/situations/covid-19
https://www.nature.com/articles/s41580-021-00418-x#citeas
https://www.science.org/doi/10.1126/science.abd9909
Zhang, Haoran et al. Advances in developing ACE2 derivatives against SARS-CoV-2. The Lancet Microbe, Volume 4, Issue 5, e369 - e378 (2023). https://doi.org/10.1016/S2666-5247(23)00011-3
PDB Files
[1]https://www.rcsb.org/structure/7UHB
Student Contributors
- Giavanna Yowell
- Shea Bailey
- Matthew Pereira