User:Reesha Bhagat/Sandbox 1
From Proteopedia
< User:Reesha Bhagat(Difference between revisions)
| (3 intermediate revisions not shown.) | |||
| Line 25: | Line 25: | ||
====Alpha-Helix Bundle==== | ====Alpha-Helix Bundle==== | ||
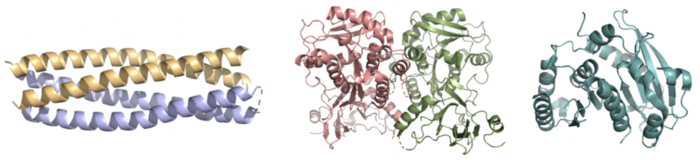
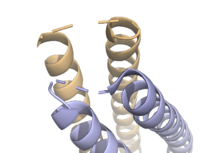
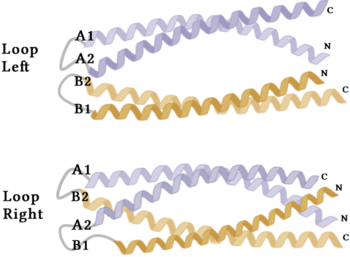
| - | The crystallization of the ''de novo'' protein, WA20, revealed | + | The crystallization of the ''de novo'' protein, WA20, revealed an <scene name='10/1075192/Overlay_rotate/3'>alpha-helix bundle</scene> homodimer structure, which served as the foundation for the design of Syn-F4 <ref name="Arai"/>. While structurally similar, Syn-F4 features a novel active site characterized by the presence of a <scene name='10/1075192/Central_hole_lone/2'>central hole</scene> <ref name="Kurihara"/>. [[Image:LOOOOPS.png|350 px|left|thumb|'''Figure 4.''' A schematic representing the differential conformations of the "loop-left" and "loop-right" Syn-F4 structures (Image created in BioRender.com).]] The symmetry of the four-helix bundle, combined with the configuration of the central hole, facilitates catalytic activity by effectively cleaving the ester bond in ferric enterobactin <ref name="Kurihara"/>. |
====Stabilizing Factors==== | ====Stabilizing Factors==== | ||
| Line 31: | Line 31: | ||
The <scene name='10/1075192/Terminal_residues/3'>terminal residues</scene> on either side of the bundles largely contribute to the stability of Syn-F4. Due to the symmetry of the chains, both '''{{Font color|#a899e6|chain A}}''' and '''{{Font color|#d6b588|chain B}}''' display the same residues and are linked by unresolved residues ('''Fig. 3''') <ref name="Kurihara"/>. The accumulated negative charge on the C-termini is stabilized by positively-charged residues (H10, H45, H46, R102), and conversely, the accumulated positive charge on the N-termini is stabilized by negatively-charged residues (D59). Additionally, the <scene name='10/1075192/H2ophobic/2'>hydrophobic residues</scene> within the core of Syn-F4 interact via Van der Waals to promote association between the helices. | The <scene name='10/1075192/Terminal_residues/3'>terminal residues</scene> on either side of the bundles largely contribute to the stability of Syn-F4. Due to the symmetry of the chains, both '''{{Font color|#a899e6|chain A}}''' and '''{{Font color|#d6b588|chain B}}''' display the same residues and are linked by unresolved residues ('''Fig. 3''') <ref name="Kurihara"/>. The accumulated negative charge on the C-termini is stabilized by positively-charged residues (H10, H45, H46, R102), and conversely, the accumulated positive charge on the N-termini is stabilized by negatively-charged residues (D59). Additionally, the <scene name='10/1075192/H2ophobic/2'>hydrophobic residues</scene> within the core of Syn-F4 interact via Van der Waals to promote association between the helices. | ||
| - | The orientation of the alpha helices and their associated interactions could not be definitively determined due to the absence of loop residues in the crystallized structure of Syn-F4 <ref name="Kurihara"/>. To address this, molecular dynamics (MD) simulations were performed to estimate the predominant conformation of the helices <ref name="Kurihara"/>. Two potential conformations were identified: "loop left" and "loop right" ('''Fig. 4''') <ref name="Kurihara"/>. Among these, the "loop left" conformation demonstrated greater stability and was therefore considered the preferred structure ('''Fig. 4''') <ref name="Kurihara"/>. | + | The orientation of the alpha helices and their associated interactions could not be definitively determined due to the absence of loop residues in the crystallized structure of Syn-F4 ('''Fig. 3''') <ref name="Kurihara"/>. To address this, molecular dynamics (MD) simulations were performed to estimate the predominant conformation of the helices <ref name="Kurihara"/>. Two potential conformations were identified: "loop left" and "loop right" ('''Fig. 4''') <ref name="Kurihara"/>. Among these, the "loop left" conformation demonstrated greater stability and was therefore considered the preferred structure ('''Fig. 4''') <ref name="Kurihara"/>. |
== Function == | == Function == | ||
| Line 39: | Line 39: | ||
====Active Site==== | ====Active Site==== | ||
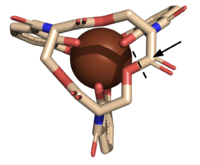
| - | The <scene name='10/1075192/Active_site/5'>active site</scene> of Syn-F4 lies within the central hole of the structure, made up of five catalytic residues (E26, H74, R77, K78, and R85) that facilitate binding to ferric enterobactin and catalyze hydrolysis of the scissile (ester) bond, which is indicated by the dotted line ('''Fig. 5''') <ref name="Kurihara"/>. The mutation of any of these five | + | The <scene name='10/1075192/Active_site/5'>active site</scene> of Syn-F4 lies within the central hole of the structure, made up of five catalytic residues (E26, H74, R77, K78, and R85) that facilitate binding to ferric enterobactin and catalyze hydrolysis of the scissile (ester) bond, which is indicated by the dotted line ('''Fig. 5''') <ref name="Kurihara"/>. The mutation of any of these five residues completely abrogates the function of the enzyme <ref name="Kurihara"/>. |
====Mechanism==== | ====Mechanism==== | ||
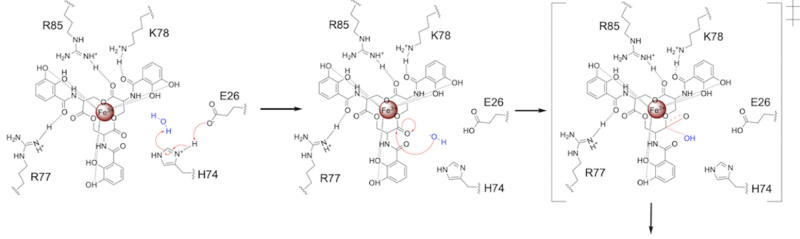
| - | The mechanism of Syn-F4 was proposed to function at pH 6.0 via a catalytic dyad with histidine (H74) and glutamate (E26) residues and water attacking as the nucleophile <ref name="Kurihara"/>. The electrophile is | + | The mechanism of Syn-F4 was proposed to function at pH 6.0 via a catalytic dyad with histidine (H74) and glutamate (E26) residues and water attacking as the nucleophile <ref name="Kurihara"/>. The electrophile is a carbonyl carbon involved in one of the ester bonds of the ligand, denoted by an arrow ('''Fig. 5'''). A representation of the possible mechanism of Syn-F4 performing hydrolysis of the scissile bond of ferric enterobactin follows: |
=====Step 1===== | =====Step 1===== | ||
| - | E26 first deprotonates H74, which then deprotonates | + | E26 first deprotonates H74, which then deprotonates H<sub>2</sub>O in turn ('''Fig. 6'''). The newly-formed hydroxide attacks the electrophile, kicking electron density up onto the carbonyl oxygen in the first transition state ('''Fig. 6'''). |
| - | [[Image: SynMech1.png|800 px|center|thumb|'''Figure 6.''' Step 1. Activation of water as the nucleophile and attack of the | + | [[Image: SynMech1.png|800 px|center|thumb|'''Figure 6.''' Step 1. Activation of water as the nucleophile and attack of the carbonyl electrophile. The first transition state is depicted.]] |
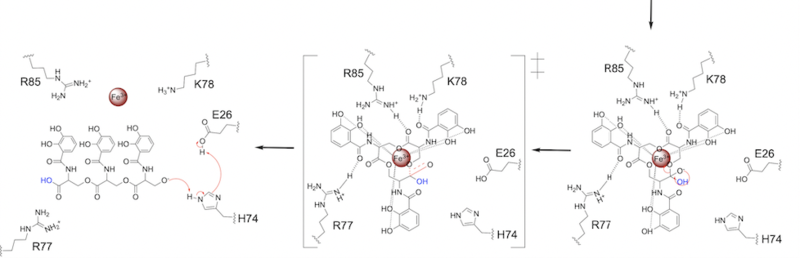
=====Step 2===== | =====Step 2===== | ||
| - | The second transition state is characterized by the electron density on the | + | The second transition state is characterized by the electron density on the carbonyl oxygen collapsing and kicking off the alcohol leaving group ('''Fig. 7'''). As the ferric enterobactin structure linearizes, Fe<sup>3+</sup> is released, and the enzyme is reset by the re-protonation of H74 by E26 ('''Fig. 7'''). |
[[Image: SynMech22222.png|800 px|center|thumb|'''Figure 7.''' Step 2. The second transition state and linearization of ferric enterobactin are depicted. Iron is released and the enzyme resets itself.]] | [[Image: SynMech22222.png|800 px|center|thumb|'''Figure 7.''' Step 2. The second transition state and linearization of ferric enterobactin are depicted. Iron is released and the enzyme resets itself.]] | ||
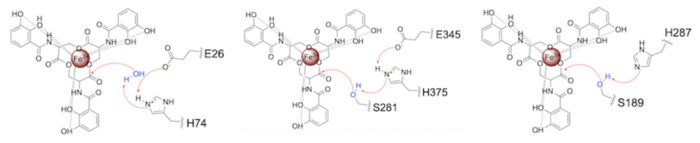
| - | ====Related | + | ====Related Enzymes==== |
Both the structure and mechanism of Syn-F4 differ from other native ferric enterobactin esterases whose structures have been previously determined. Native Fes and IroE enzymes are both serine hydrolases, which means a serine residue acts as their nucleophile <ref name="Lin"/> <ref name="Larsen"/> <ref name="Timofeeva">PMID:36432794</ref>. Native <scene name='10/1075192/Fes_residues/11'>Fes</scene> is a homodimer that acts via a catalytic triad (S281, E345, and H375), whereas <scene name='10/1075192/Iroe_residue/6'>IroE</scene> acts via a catalytic dyad (S189 and H287), similar to Syn-F4 ('''Fig. 8''') <ref name="Lin"/> <ref name="Larsen"/> <ref name="Timofeeva"/>. | Both the structure and mechanism of Syn-F4 differ from other native ferric enterobactin esterases whose structures have been previously determined. Native Fes and IroE enzymes are both serine hydrolases, which means a serine residue acts as their nucleophile <ref name="Lin"/> <ref name="Larsen"/> <ref name="Timofeeva">PMID:36432794</ref>. Native <scene name='10/1075192/Fes_residues/11'>Fes</scene> is a homodimer that acts via a catalytic triad (S281, E345, and H375), whereas <scene name='10/1075192/Iroe_residue/6'>IroE</scene> acts via a catalytic dyad (S189 and H287), similar to Syn-F4 ('''Fig. 8''') <ref name="Lin"/> <ref name="Larsen"/> <ref name="Timofeeva"/>. | ||
| Line 74: | Line 74: | ||
==Student Contributors== | ==Student Contributors== | ||
| - | *Milica Nenadovich | ||
*Reesha Bhagat | *Reesha Bhagat | ||
| + | *Milica Nenadovich | ||
Current revision
Syn-F4, a de novo Ferric Enterobactin Esterase
| |||||||||||
Student Contributors
- Reesha Bhagat
- Milica Nenadovich