Sandbox 326
From Proteopedia
| (7 intermediate revisions not shown.) | |||
| Line 2: | Line 2: | ||
| - | 3B7F is a currently unknown protein in terms of its function. Based on current structural analysis, it consists of one unique chain with a mass of | + | 3B7F is a currently unknown protein in terms of its function. Based on current structural analysis, it consists of one unique chain with a mass of 43.34 kDa and an atom count of 3,216. The mass was found by multiply the 394 amino acids of the protein by the mass of each amino acid (~0.110 kDa). Based on previous studies, 3B7F is assumed to be a glycosyl hydrolase, however, the function is still not entirely known.[1] Through the following procedures and data collection, the goal of this research was to analyze the sequence and structure of 3B7F in order to better understand its enzymatic function. |
'''Research Question:''' What is the function of the 3B7F protein, and how can this be determined through both computational and wet lab techniques? | '''Research Question:''' What is the function of the 3B7F protein, and how can this be determined through both computational and wet lab techniques? | ||
| - | '''Relevance:''' The goal of this research is to determine the function of the 3BF7 protein in order to evaluate whether it can degrade xyloglucan or other types of carbohydrates and glycoconjugates in plants. Knowing this will allow future researchers to be able to better understand xyloglucan/carbohydrate glycoconjugate degradation in plants, and allow for known pathways to be expanded upon. Through this experimentation, we can also learn more about the ways in which both computational bioinformatics and wet lab techniques can aid in determining the function of a protein with a known structure. | + | '''Relevance:''' The goal of this research is to determine the function of the 3BF7 protein in order to evaluate whether it can degrade xyloglucan or other types of carbohydrates and glycoconjugates in plants. Knowing this will allow future researchers to be able to better understand xyloglucan/carbohydrate/glycoconjugate degradation in plants, and allow for known pathways to be expanded upon. Through this experimentation, we can also learn more about the ways in which both computational bioinformatics and wet lab techniques can aid in determining the function of a protein with a known structure. |
'''Hypothesis:''' 3B7F is a xyloglucanase, a type of glycosyl hydrolase that acts to degrade xyloglucan in plant cell walls. It presents optimal activity in fairly acidic conditions and demonstrates potentially satisfactory binding with PNP phosphate and lysine p nitroanilide. | '''Hypothesis:''' 3B7F is a xyloglucanase, a type of glycosyl hydrolase that acts to degrade xyloglucan in plant cell walls. It presents optimal activity in fairly acidic conditions and demonstrates potentially satisfactory binding with PNP phosphate and lysine p nitroanilide. | ||
| - | <StructureSection load='3B7F' size='400' side='right' caption='Structure of 3B7F' scene=''> | + | <StructureSection load='3B7F' size='400' side='right' caption='Secondary Structure of 3B7F' scene=''> |
== Methods == | == Methods == | ||
| - | '''SPRITE: | + | '''SPRITE:''' |
| - | ''' | + | |
The PBD ID for the protein of interest (3B7F) was entered and 2-residue hits excluded. Once the search was complete, "List of Hits" results were obtained. "Full details" result alignments also analyzed. Hits by each side of protein viewed by clicking "Arranged by sites" function. Alignments with an RMSD below 2.0 Angstroms were reviewed. | The PBD ID for the protein of interest (3B7F) was entered and 2-residue hits excluded. Once the search was complete, "List of Hits" results were obtained. "Full details" result alignments also analyzed. Hits by each side of protein viewed by clicking "Arranged by sites" function. Alignments with an RMSD below 2.0 Angstroms were reviewed. | ||
| Line 76: | Line 76: | ||
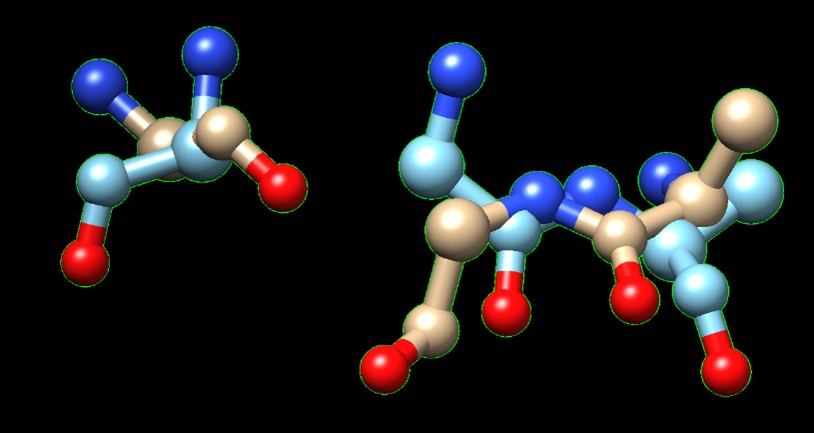
Based on the findings through SPRITE and Chimera, 1XNY_C00 had the lowest RMSD value at 1.875 angstroms. Therefore, it is hypothesized that 3B7F was a carboxylase. | Based on the findings through SPRITE and Chimera, 1XNY_C00 had the lowest RMSD value at 1.875 angstroms. Therefore, it is hypothesized that 3B7F was a carboxylase. | ||
| - | [[Image:SPRITE.jpg]] | + | [[Image:SPRITE.jpg]] [[Image:Chimera.jpg]] |
| + | A 183 GLY matches A 207 GLY/ | ||
| + | B 419 GLY matches A 154 GLY/ | ||
| + | B 420 ALA matches A 153 ALA | ||
| - | Figure 1. SPRITE and Chimera binding of 3B7F (yellow) with 1XNY_C00 (green). | + | '''Figure 1.''' SPRITE and Chimera binding of 3B7F (yellow) with 1XNY_C00 (green). |
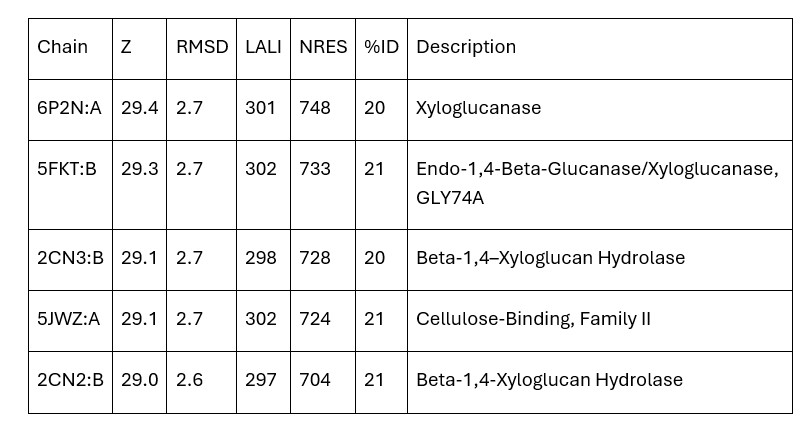
| - | + | Dali provided mainly xyloglucanase matches and no matches were carboxylases. Therefore, the initial hypothesis that 3B7F was a carboxylase was no longer supported. Based on the Dali results, it is now hypothesized that 3B7F is a xyloglucanase. [2] | |
| - | + | '''Table 1.''' Top Matches on Dali (Z: greater than 4.0 indicates structural significance; RMSD: root mean square deviation (distance in Angstroms between superimposed molecules); LALI: length of alignment; NRES: number of residues in hit structure; %ID: similarity between hit sequence and query sequence). | |
| + | [[Image:TableDali.jpg]] | ||
| - | + | [[Image:BLAST.jpg]] | |
| + | '''Figure 2.''' Superimposition of 6P2N:A (orange) and 3B7F (green). | ||
| - | <table> | ||
| - | <tr> | ||
| - | <td> Chain </td> | ||
| - | <td> Z </td> | ||
| - | <td> RMSD </td> | ||
| - | <td> LALI </td> | ||
| - | <td> NRES </td> | ||
| - | <td> %ID </td> | ||
| - | <td> Description </td> | ||
| - | </tr> | ||
| - | + | Xyloglucanases break down xyloglucan, a hemicellulose in plant cell walls. The substrate is xyloglucan and water for the (endo-)beta-1,4-xyloglucanases, and a common cofactor is calcium. [3] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The BLAST search shows glycosyl hydrolases having similar sequences to 3B7F. The xyloglucanases/glycosyl hydrolases from the BLAST search come from Cupriavidus, a type of gram-negative bacteria. The E-values were 0.0 for most of the results, signifying high matches. The scores were high as well, with percent identification in the 90 percentage range. The query coverage was also 100% for most of the results. Based on the results, the protein family of 3B7F is likely sialidase, which breaks down sialic acid into carbohydrates. Sialidases are a family of proteins that include glycosyl hydrolases and xyloglucanases, which further supports the hypothesis that 3B7F is a xyloglucanase. [4] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Using InterPro to Predict Protein Function == | == Using InterPro to Predict Protein Function == | ||
| - | The family with the closest similarities to 3B7F is xyloglucanase, which matches the BLAST and Dali findings. These enzymes act on xyloglucan, a plant wall polysaccharide, by breaking the glucosidic bonds of unbranched glucose residues. Most of the proteins in this family bind to cell wall structures or participate in photosystem II, which correlates with the expected activity of 3B7F, since it would be acting on plant cell walls. This family consists mainly of bacteria, as well as eukaryotes and archaea. This also matches the BLAST data because it suggested that the sequence was commonly found in bacteria. The proteomes are a lot of bacteria and marine life. The most closely matched structures are different xyloglucanases and oligoxyloglucan reducing end-specific cellobiohydralases, which is the superfamily that the protein belongs to. The main pathways are also structural degradation pathways, which also aligns with the results we have gained so far. | + | The family with the closest similarities to 3B7F is xyloglucanase, which matches the BLAST and Dali findings. These enzymes act on xyloglucan, a plant wall polysaccharide, by breaking the glucosidic bonds of unbranched glucose residues. Most of the proteins in this family bind to cell wall structures or participate in photosystem II, which correlates with the expected activity of 3B7F, since it would be acting on plant cell walls. This family consists mainly of bacteria, as well as eukaryotes and archaea. This also matches the BLAST data because it suggested that the sequence was commonly found in bacteria. The proteomes are a lot of bacteria and marine life. The most closely matched structures are different xyloglucanases and oligoxyloglucan reducing end-specific cellobiohydralases, which is the superfamily that the protein belongs to. The main pathways are also structural degradation pathways, which also aligns with the results we have gained so far. [5] |
== Molecular Docking with SwissDock == | == Molecular Docking with SwissDock == | ||
| - | Based on the hydrolase ligands that we analyzed, PNP phosphate seems to be the best ligand for 3B7F, with the highest binding affinity of -8.189 kcal/mol, and the lowest binding affinity of -6.146 kcal/mol. This is a very strong binding affinity, so this substrate was used for protein assays. Lysine p nitroanilide also has its highest binding affinity at -7.286 kcal/mol and | + | Based on the hydrolase ligands that we analyzed, PNP phosphate seems to be the best ligand for 3B7F, with the highest binding affinity of -8.189 kcal/mol, and the lowest binding affinity of -6.146 kcal/mol. This is a very strong binding affinity, so this substrate was used for protein assays. Lysine p nitroanilide also has its highest binding affinity at -7.286 kcal/mol and the lowest binding affinity of -6.551 kcal/mol, indicating that it is another suitable substrate for 3B7F. [6] |
| - | [[Image: | + | [[Image:SwissDock1.jpg]] |
| + | '''Figure 3.''' 3B7F Binding with PNPP at -8.189 kcal/mol. | ||
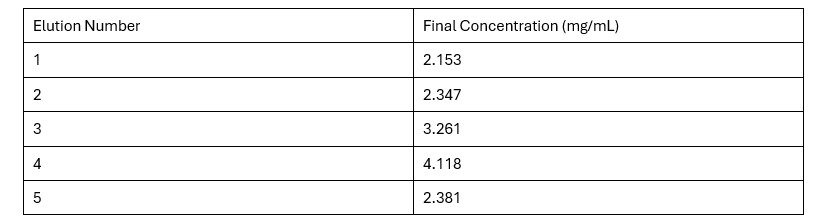
| - | + | == Protein Concentration == | |
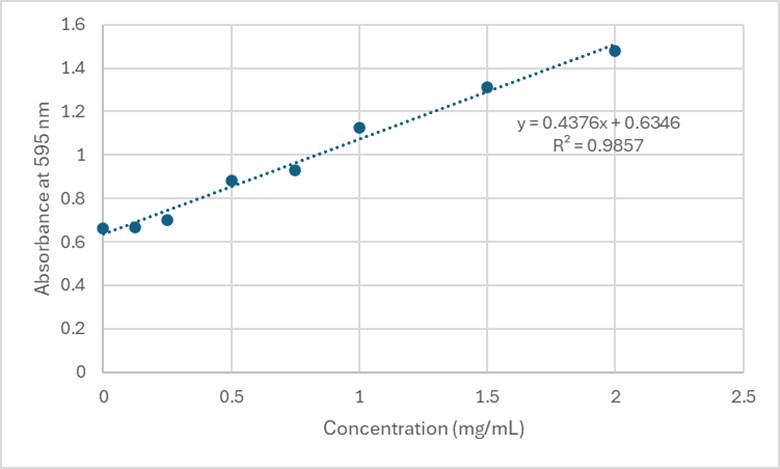
| - | + | Based on the Bradford Assay, elution 4 had the most protein (4.118 mg/mL), while elution 3 had the second most amount of protein (3.261 mg/mL). The rest of the elutions had protein concentrations around 2 mg/mL. The R² value of the standard curve was 0.9857, indicating fairly accurate results. | |
| - | + | [[Image:StandardCurve.jpg]] | |
| + | '''Figure 4.''' Standard Curve at 595 nm for 3B7F Elutions. | ||
| - | + | '''Table 2.''' Final Protein Concentration of Each Elution | |
| + | [[Image:FinalConc.jpg]] | ||
| - | + | == Protein Analysis == | |
| - | + | Possibly due to errors during purification, no bands were seen on the SDS-PAGE gel for 3B7F. | |
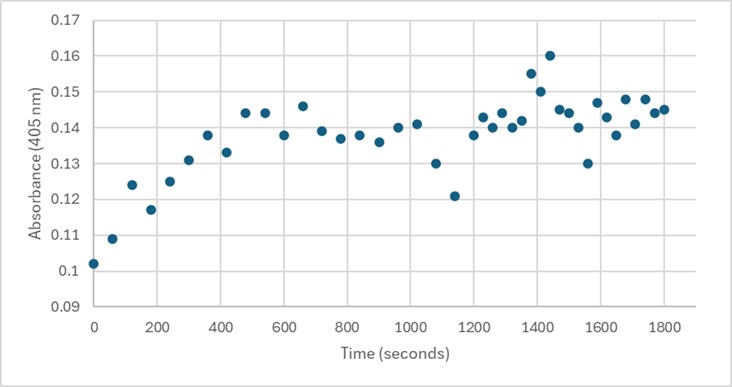
| - | + | Several conditions were analyzed through enzyme activity assays. Initially, 50 uL of 3B7F elution 4 with 1 mg/mL PNPP demonstrated some linearity between 0-600 seconds. However, this was not able to be replicated. Both 25 uL and 75 uL of 3B7F elution 4 were also used with PNPP, and no linearity was found. Due to the enzyme’s optimal conditions, it was also cooled down in -20°C for five minutes, however, no linearity was seen. The assay was also perfomed with elution 1 to identify if any protein was present after the wash. Again, no linearity was shown. PNPA was also used as a substrate with elution 4, and pHs of 1.5 and 5.0 were also used during separate trials in order to identify the optimal conditions of the enzyme. [7] However, these assays still showed no linearity. It was determined that there was an error during purification, leading to unidentifiable concentrations of 3B7F for enzymatic study. | |
| - | + | [[Image:Assay.jpg]] | |
| + | '''Figure 5.''' 50 uL of 3B7F Elution 4 with 1 mg/mL PNPP. | ||
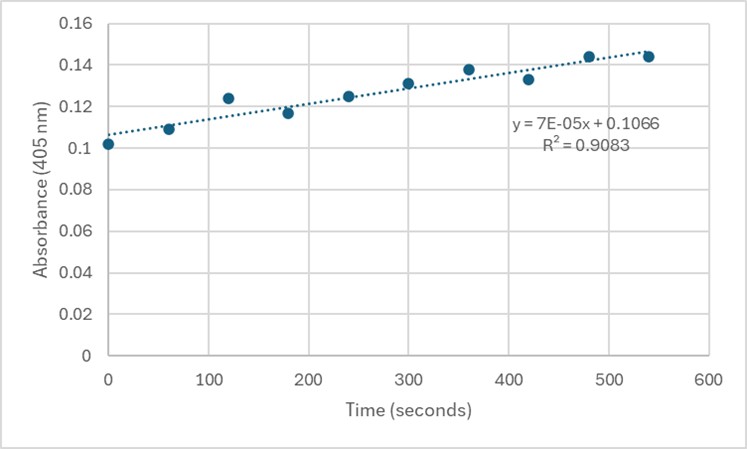
| - | + | [[Image:Assay2.jpg]] | |
| + | Slope = 7 x 10⁻⁵ Abs/sec | ||
| + | C = 7 x 10⁻⁵/18000*1 | ||
| + | = 3.89 x 10⁻⁹ M/sec | ||
| - | Figure | + | '''Figure 6.''' Zoom in with linear trendline of Figure 5 and enzyme activity calculations. |
| - | + | == Conclusion == | |
| + | Based on the results, no enzymatic function of 3B7F can be appropriately determined. However, based on the computational results, it is likely that 3B7F has xyloglucanase activity. In the future, the protein should be grown again in order to retest the protein’s enzymatic activity after a proper purification. The results that were successfully gathered did show accuracy and precision. However, likely due to incorrect solution making, the protein was not able to be properly purified, and therefore, was not present in the elutions. Interestingly enough, the Bradford Assay did show protein concentrations, but they likely were not for the desired protein (3B7F), which may be the reason for no protein present at the expected mass of approximately 43 kDa. If 3B7F is confirmed as a xyloglucanase, it can possibly be used for drug targeting in the colon through xyloglucan coatings. [8] | ||
| + | </StructureSection> | ||
| + | == References == | ||
| - | + | [1] RCSB PDB - 3B7F: Crystal structure of a putative glycosyl hydrolase with bnr repeats (reut_b4987) from ralstonia eutropha jmp134 at 2.20 A resolution. Rcsb.org. https://www.rcsb.org/structure/3b7f (accessed 2025-03-09). | |
| - | + | [2] Dali server. ekhidna2.biocenter.helsinki.fi. http://ekhidna2.biocenter.helsinki.fi/dali/. | |
| - | + | [3] Glycoside hydrolases - CAZypedia. www.cazypedia.org. https://www.cazypedia.org/index.php/Glycoside_hydrolases. | |
| - | + | [4] NCBI. BLAST: Basic Local Alignment Search Tool. Nih.gov. https://blast.ncbi.nlm.nih.gov/Blast.cgi. | |
| - | + | [5] InterPro EMBL-EBI. InterPro protein sequence analysis & classification < InterPro < EMBL-EBI. Ebi.ac.uk. https://www.ebi.ac.uk/interpro/. | |
| - | + | [6] SwissDock. www.swissdock.ch. https://www.swissdock.ch/. | |
| - | + | [7] Shi, H.; Guo, J.; Yan, X.; Cui, G.; Tan, Z.; Zhu, X.; Zhou, J.; He, S.; Wang, T.; Li, X. Characterization of a Xyloglucananse in Biodegradation of Woody Plant Xyloglucan from Caldicellulosiruptor Kronotskyensis. BioResources 2021, 17 (1), 673–681. https://doi.org/10.15376/biores.17.1.673-681. | |
| - | + | [8] Doggwiler, V.; Lanz, M.; Paredes, V.; Lipps, G.; Imanidis, G. Tablet Formulation with Dual Control Concept for Efficient Colonic Drug Delivery. International Journal of Pharmaceutics 2023, 631, 122499. https://doi.org/10.1016/j.ijpharm.2022.122499. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<references/> | <references/> | ||
Current revision
Characterization and Preliminary Functionality of 3B7F
3B7F is a currently unknown protein in terms of its function. Based on current structural analysis, it consists of one unique chain with a mass of 43.34 kDa and an atom count of 3,216. The mass was found by multiply the 394 amino acids of the protein by the mass of each amino acid (~0.110 kDa). Based on previous studies, 3B7F is assumed to be a glycosyl hydrolase, however, the function is still not entirely known.[1] Through the following procedures and data collection, the goal of this research was to analyze the sequence and structure of 3B7F in order to better understand its enzymatic function.
Research Question: What is the function of the 3B7F protein, and how can this be determined through both computational and wet lab techniques?
Relevance: The goal of this research is to determine the function of the 3BF7 protein in order to evaluate whether it can degrade xyloglucan or other types of carbohydrates and glycoconjugates in plants. Knowing this will allow future researchers to be able to better understand xyloglucan/carbohydrate/glycoconjugate degradation in plants, and allow for known pathways to be expanded upon. Through this experimentation, we can also learn more about the ways in which both computational bioinformatics and wet lab techniques can aid in determining the function of a protein with a known structure.
Hypothesis: 3B7F is a xyloglucanase, a type of glycosyl hydrolase that acts to degrade xyloglucan in plant cell walls. It presents optimal activity in fairly acidic conditions and demonstrates potentially satisfactory binding with PNP phosphate and lysine p nitroanilide.
| |||||||||||
References
[1] RCSB PDB - 3B7F: Crystal structure of a putative glycosyl hydrolase with bnr repeats (reut_b4987) from ralstonia eutropha jmp134 at 2.20 A resolution. Rcsb.org. https://www.rcsb.org/structure/3b7f (accessed 2025-03-09).
[2] Dali server. ekhidna2.biocenter.helsinki.fi. http://ekhidna2.biocenter.helsinki.fi/dali/.
[3] Glycoside hydrolases - CAZypedia. www.cazypedia.org. https://www.cazypedia.org/index.php/Glycoside_hydrolases.
[4] NCBI. BLAST: Basic Local Alignment Search Tool. Nih.gov. https://blast.ncbi.nlm.nih.gov/Blast.cgi.
[5] InterPro EMBL-EBI. InterPro protein sequence analysis & classification < InterPro < EMBL-EBI. Ebi.ac.uk. https://www.ebi.ac.uk/interpro/.
[6] SwissDock. www.swissdock.ch. https://www.swissdock.ch/.
[7] Shi, H.; Guo, J.; Yan, X.; Cui, G.; Tan, Z.; Zhu, X.; Zhou, J.; He, S.; Wang, T.; Li, X. Characterization of a Xyloglucananse in Biodegradation of Woody Plant Xyloglucan from Caldicellulosiruptor Kronotskyensis. BioResources 2021, 17 (1), 673–681. https://doi.org/10.15376/biores.17.1.673-681.
[8] Doggwiler, V.; Lanz, M.; Paredes, V.; Lipps, G.; Imanidis, G. Tablet Formulation with Dual Control Concept for Efficient Colonic Drug Delivery. International Journal of Pharmaceutics 2023, 631, 122499. https://doi.org/10.1016/j.ijpharm.2022.122499.