We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox3H04
From Proteopedia
(Difference between revisions)
(Undo revision 4332105 by Cameron Snelbaker (Talk)) |
|||
| (20 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | ==Overview== | ||
| - | <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | This is a default text for your page '''Sandbox3H04'''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
'''General Structure and Origins''' | '''General Structure and Origins''' | ||
| Line 12: | Line 8: | ||
== Structure/Sequence Analysis == | == Structure/Sequence Analysis == | ||
| - | + | <StructureSection load='3H04' size='400' side='right' caption='PDB Structure of 3H04' scene=''> | |
'''Sequence Analysis''' | '''Sequence Analysis''' | ||
| Line 20: | Line 16: | ||
'''Chimera''' | '''Chimera''' | ||
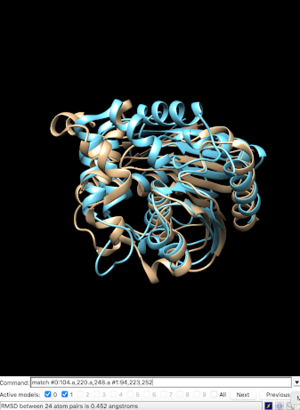
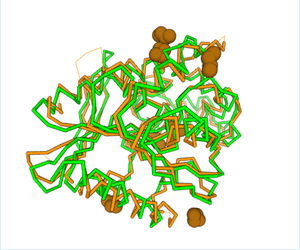
| - | The structural viewing program Chimera was used to view the specific amino acids from SPRITE in the theorized functional group. 1a8q was the primary match used to model matching | + | The structural viewing program Chimera was used to view the specific amino acids from SPRITE in the theorized functional group. 1a8q was the primary match used to model matching 3H04’s 104 serine at 1a8q’s 94, 220 aspartate at 223, and 248 histidine at 252 respectively with a RMSD of 0.452. An overlap was performed to view the functional alignment which can be viewed here.[[Image:ChimeraAlignmentBS.png |300px|center|thumb|]] |
'''BLAST''' | '''BLAST''' | ||
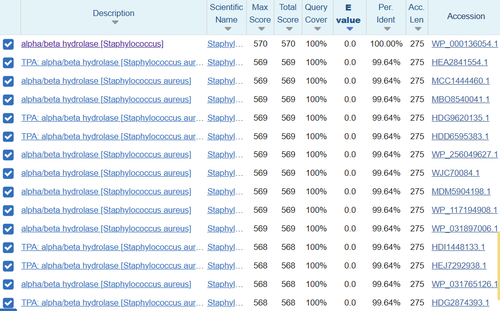
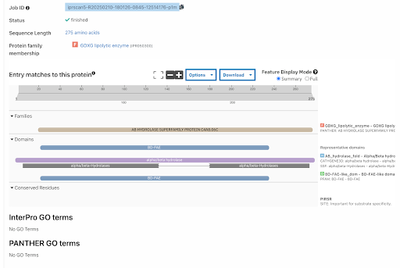
| - | BLAST takes the exact amino acid chain of the protein and runs against a database to view similar sequences which may have similar function. When the chain was inputted into the BLAST software, the results were measured against a “score” showing the significant alignments as well as an E-score which helps show how good of a match it is. The one with the lowest E-score was E. coli MutT with a score of 3 x 10-88, showing that it is a great match. The total score was at the max value with 100% query coverage meaning the sequence was identical. The next several resulting sequences all belonged to Escherichia coli with extremely small E-values and nearly 100% coverage against the 3H04 sequence. The results came from the preliminary search with the MutT line for sequence, however another query was run based upon the PDB sequence which should have more specific results. Again the Escherichia coli appeared with incredible results for Chain A, Nucleoside Triphosphate pyrophosphohydrolase. A third query was used from the RCSB chain for 3H04 “MTEIKYKVITKDAFALPYTIIKAKNQPTKGVIVYIHGGGLMFGKANDLSPQYIDILTEHYDLIQLSYRLLPEVSLDCIIEDVYASFDAIQSQYSNCPIFTFGRSSGAYLSLLIARDRDIDG-VIDFYGYSRINTEPFKTTNSYYAKIAQSINETMIAQLTSPTPVVQDQIAQRFLIYVYARGTGKWINMINIADYTDSKYNIAPDELKTLPPVFIAHC-NGDYDVPVEESEHIMNHVPHSTFERVNKNEHDFDRRPNDEAITIYRKVVDFLNAITMV” which received a top hit of alpha/beta hydrolase [Staphylococcus] with a max score of 570, query coverage of 100%, and E-value of approximately 0.0. The result was incredible because it showed this is exactly where 3H04 arose based on the E-value and gave solid evidence that alpha/beta hydrolase was the function of 3H04. | + | |
| + | BLAST takes the exact amino acid chain of the protein and runs against a database to view similar sequences which may have similar function. When the chain was inputted into the BLAST software, the results were measured against a “score” showing the significant alignments as well as an E-score which helps show how good of a match it is. The one with the lowest E-score was E. coli MutT with a score of 3 x 10-88, showing that it is a great match. The total score was at the max value with 100% query coverage meaning the sequence was identical. The next several resulting sequences all belonged to Escherichia coli with extremely small E-values and nearly 100% coverage against the 3H04 sequence. The results came from the preliminary search with the MutT line for sequence, however another query was run based upon the PDB sequence which should have more specific results. Again the Escherichia coli appeared with incredible results for Chain A, Nucleoside Triphosphate pyrophosphohydrolase. A third query was used from the RCSB chain for 3H04 “MTEIKYKVITKDAFALPYTIIKAKNQPTKGVIVYIHGGGLMFGKANDLSPQYIDILTEHYDLIQLSYRLLPEVSLDCIIEDVYASFDAIQSQYSNCPIFTFGRSSGAYLSLLIARDRDIDG-VIDFYGYSRINTEPFKTTNSYYAKIAQSINETMIAQLTSPTPVVQDQIAQRFLIYVYARGTGKWINMINIADYTDSKYNIAPDELKTLPPVFIAHC-NGDYDVPVEESEHIMNHVPHSTFERVNKNEHDFDRRPNDEAITIYRKVVDFLNAITMV” which received a top hit of alpha/beta hydrolase [Staphylococcus] with a max score of 570, query coverage of 100%, and E-value of approximately 0.0. The result was incredible because it showed this is exactly where 3H04 arose based on the E-value and gave solid evidence that alpha/beta hydrolase was the function of 3H04.[[Image:INTERPRO.png |500px|center|thumb|]] | ||
'''Structure Analysis''' | '''Structure Analysis''' | ||
| Line 35: | Line 32: | ||
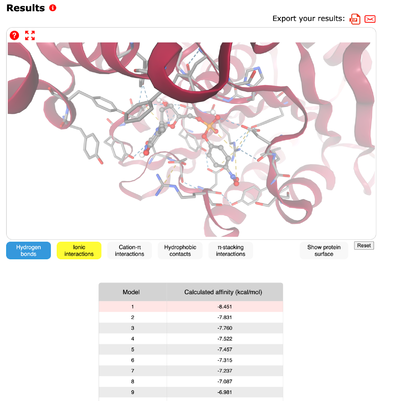
The SwissDock software enables the visualization of what ligands would fit best with the structure of the protein. This is determined by looking at the “calculated affinity” for each of the ligands in a static setting. The strongest match for 3H04 was p-Nitrophenyl thymidine-5'-monophosphate with a very high affinity of -8.451 kcal/mol. Other matches occurred with leucine P-nitro with an affinity of -6.505 kcal/mol, mannobiose with -6.669 kcal/mol, PNP Alpha D Glucopyranoside with -7.089 kcal/mol, and PNP N Acetyl beta D glucosaminide with -6.955 kcal/mol. All results showed similar structures with the ability to be cleaved. | The SwissDock software enables the visualization of what ligands would fit best with the structure of the protein. This is determined by looking at the “calculated affinity” for each of the ligands in a static setting. The strongest match for 3H04 was p-Nitrophenyl thymidine-5'-monophosphate with a very high affinity of -8.451 kcal/mol. Other matches occurred with leucine P-nitro with an affinity of -6.505 kcal/mol, mannobiose with -6.669 kcal/mol, PNP Alpha D Glucopyranoside with -7.089 kcal/mol, and PNP N Acetyl beta D glucosaminide with -6.955 kcal/mol. All results showed similar structures with the ability to be cleaved. | ||
| + | [[Image:swissdik.png|400px|center|thumb|]] | ||
== Proposed Functionality == | == Proposed Functionality == | ||
The proposed functionality is based upon multiple computer-based analysis and experimental data. There were several different matches overall, however all of them had the same general function of alpha-beta hydrolase. The connection to 3H04 and alpha-beta hydrolase will be explored more in the following sections. | The proposed functionality is based upon multiple computer-based analysis and experimental data. There were several different matches overall, however all of them had the same general function of alpha-beta hydrolase. The connection to 3H04 and alpha-beta hydrolase will be explored more in the following sections. | ||
| Line 48: | Line 46: | ||
'''Overview''' | '''Overview''' | ||
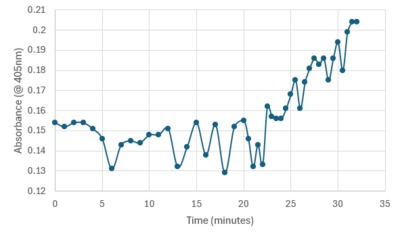
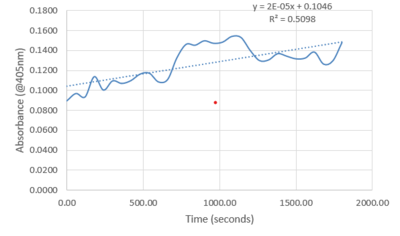
| - | Protein Purification was the first step in getting to a point of experimental data. The protein was grown in E. coli solution that had been tagged with an antibiotic resistance to isolate the protein. The pellet was then lysed with a buffer and sonicated to break up the sample. The resulting sample was centrifuged and the liquid was collected and placed into a separatory column to gather elutions. All steps had portions that were taken throughout to view where the protein was best concentrated. The first experimental occurred from a Bradford assay of known standards to determine which elution had the highest concentration of protein. (Graph to show elution 1 had highest amount). The second experiment was running the portions through all steps on an SDS-PAGE gel to view the purify and amount of protein. It was determined that the protein had expelled through the wash before the elutions were collected. (Include sds-page here) The wash was used to test enzymatic activity by adding PNPP, a buffer, and protein, then recording the absorbance for around 30 minutes. Testing showed a sharp increase in absorbance from 22 to 32 minutes meaning PNPP was cleaved to produce the increase in intensity. The experiment was run again at body temperature using a well plate reader with similar results but much higher increase in intensity. | + | |
| + | Protein Purification was the first step in getting to a point of experimental data. The protein was grown in E. coli solution that had been tagged with an antibiotic resistance to isolate the protein. The pellet was then lysed with a buffer and sonicated to break up the sample. The resulting sample was centrifuged and the liquid was collected and placed into a separatory column to gather elutions. All steps had portions that were taken throughout to view where the protein was best concentrated. The first experimental occurred from a Bradford assay of known standards to determine which elution had the highest concentration of protein. (Graph to show elution 1 had highest amount). The second experiment was running the portions through all steps on an SDS-PAGE gel to view the purify and amount of protein. It was determined that the protein had expelled through the wash before the elutions were collected. (Include sds-page here) The wash was used to test enzymatic activity by adding PNPP, a buffer, and protein, then recording the absorbance for around 30 minutes. Testing showed a sharp increase in absorbance from 22 to 32 minutes meaning PNPP was cleaved to produce the increase in intensity. The resulting plot is shown below. | ||
| + | [[Image:Increasingplot.png|400px|center|thumb|]] | ||
| + | The experiment was run again at body temperature using a well plate reader with similar results but much higher increase in intensity. The resulting plot is shown below. | ||
| + | [[Image:Screenshot 2025-04-27 203045.png|400px|center|thumb|]] | ||
== Relevance == | == Relevance == | ||
| Line 59: | Line 61: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | 3H04. Protein Database, 2009. https://www.rcsb.org/structure/3H04 | ||
Current revision
General Structure and Origins The total structure weighs 31.41 kDa and the atom count is 2,297. The protein 3H04 usually only occurs in bacteria, specifically staphylococcus aureus subsp. aureus Mu50. Each chain in the protein is about 275 residues long. Using InterPro, it can be seen that the protein is found in the alpha-beta hydrolase superfamily.
Family and Superfamily The family code was BD-FAE under the superfamily of alpha-beta hydrolase.
Structure/Sequence Analysis
| |||||||||||
References
3H04. Protein Database, 2009. https://www.rcsb.org/structure/3H04