We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox323
From Proteopedia
(Difference between revisions)
| (41 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | == | + | ==Structural Analysis and Proposed Functionality of 4Q7Q== |
| - | + | ||
| - | + | ||
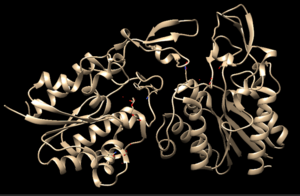
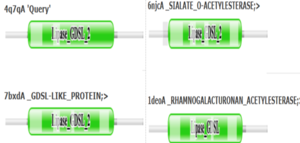
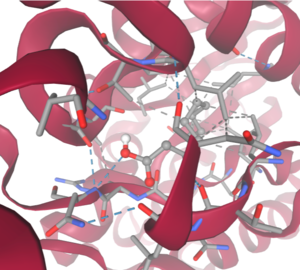
4Q7Q is a homodimeric protein complex that originates from the bacterial species Chitinophaga Pinensis and has a mass of 58.5 kDa. It is a member of the SGNH Hydrolase Superfamily with structural and sequential similarities to esterases and lipases. Current evidence suggests it causes the hydrolysis of esters and/or acetyl groups on lipids/lipid-like molecules via a catalytic triad-like active site. | 4Q7Q is a homodimeric protein complex that originates from the bacterial species Chitinophaga Pinensis and has a mass of 58.5 kDa. It is a member of the SGNH Hydrolase Superfamily with structural and sequential similarities to esterases and lipases. Current evidence suggests it causes the hydrolysis of esters and/or acetyl groups on lipids/lipid-like molecules via a catalytic triad-like active site. | ||
| Line 74: | Line 72: | ||
== Experimental Data == | == Experimental Data == | ||
| - | === | + | === Bacterium === |
| - | === Hydrolysis of Substrate === | + | Chitinophaga pinensis is an obligate aerobe, spore-forming, psychrophilic bacterium that was isolated from pine litter. Bacterium actively grows in pH 7 to 10 media with pH 7 being ideal.<ref name="BacDive">Schober, I.; Koblitz, J.; Carbasse, J. S.; Ebeling, C.; Schmidt, M. L.; Podstawka, A.; Gupta, R.; Ilangovan, V.; Chamanara, J.; Overmann, J.; Reimer, L. C. BacDive in 2025: the core database for prokaryotic strain data. Nucleic Acids Res., 2024, 53(D1), D748-D756, https://doi.org/10.1093/nar/gkae959</ref> |
| + | |||
| + | === Substrate and Hydrolysis of Substrate === | ||
| + | |||
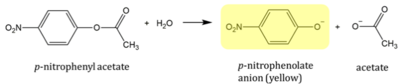
| + | p-nitrophenyl acetate (PNPA) is an aromatic compound with an ester linkage. PNPA supplied by ThermoScientific 97% pure CAS: 830-03-5. In the presence of an esterase, the compound is susceptible to cleavage. The products are acetate and p-nitrophenolate anion (PNP). PNP is yellow in color so reaction progress can be monitored by measuring the absorption peak at 405nm using a spectrophotometer.<ref name="BasilAssay">Protein Activity Assays Student Module. BASIL. | ||
| + | https://docs.google.com/document/d/1G-H0tskmSVRhC0qsDau-7-EzUcUYN8kBUHFZyBStYcs/edit?tab=t.0 (accessed 04/28/25).</ref> | ||
| + | |||
| + | [[Image:PNPA.png|400px|right|The reaction used to analyze the activity of our protein<ref name="BasilAssay">Protein Activity Assays Student Module. BASIL. | ||
| + | https://docs.google.com/document/d/1G-H0tskmSVRhC0qsDau-7-EzUcUYN8kBUHFZyBStYcs/edit?tab=t.0 (accessed 04/28/25).</ref>]] | ||
=== Spectrophotometry === | === Spectrophotometry === | ||
| - | [[Image:Ice_bath_basic_conditions_with_salt.jpg| | + | 50uL of purified protein from the first elution was added to 3mL of cold 1mg/mL PNPA. Solid sodium chloride was added to achieve a final concentration of 5mM NaCl. The absorbance peak was measured every 60 seconds for 30 minutes via a Vernier Go Direct SpectroVis Plus Spectrophotometer. Peak maximum was located at 427.8nm. The sample incubated in an ice bath between measurements. No discernible relationship between time and absorbance was noted. pH of the solution was 10.60. The instrument to measure pH was Vernier Lab Quest 3 with Tris-compatible flat pH sensor calibrated with two points. Theorizing the enzyme would increase activity at the physiological pH of common prokaryotes, we lowered the pH with 2M HCl. 2M NaOH was used to raise the pH due to the weak buffering capacity of the PNPA solution near pH 7. Final pH of the 1mg/mL PNPA solution was 7.30. |
| + | |||
| + | === Experimental Data and Discussion === | ||
| + | |||
| + | [[Image:Ice_bath_basic_conditions_with_salt.jpg|400px|left|thumb|4 degrees C reaction under basic conditions]] | ||
| + | |||
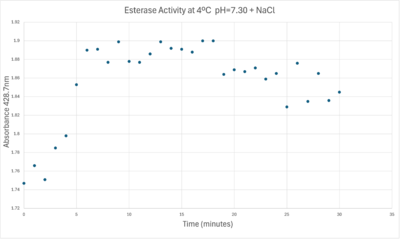
| + | 50uL of purified protein from the first elution was added to 3mL of cold 1mg/mL PNPA at pH=7.30. Chloride ion cofactor was provided by the 2M HCl. A change in absorbance at 428.7nm over time was noted. The time range of the run did not produce a linear response. The absorbance increased for the first ten minutes and plateaued. The time 0 to 10 minutes was used to create the third figure with a more refined linear range. | ||
| + | |||
| + | [[Image:Absorbancelongph7cold.png|400px|left|Absorbance at 405nm of a PNPA+Protein solution with pH = 7 and sodium chloride in a cold environment.]] | ||
| + | |||
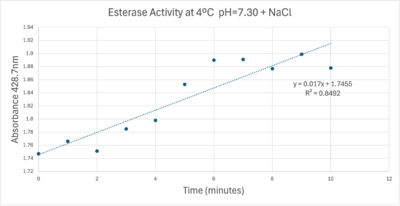
| + | [[Image:Absorbancecoldph7try.png|400px|left|A trendline showing the potential relationship between absorbance and time in a PNPA+Protein solution in a solution with a pH of 7, sodium chloride, and a external temperature of 4 C.]] | ||
| + | |||
| + | A linear equation of y=9.275x10^-7x+1x10^-4 with a coefficient of determination of 0.8492 was calculated using linear regression. | ||
| - | + | [[Image:Tablebeerlaw.png|1000px|left|thumb|Beer-Lambert Law sample calculation.]] | |
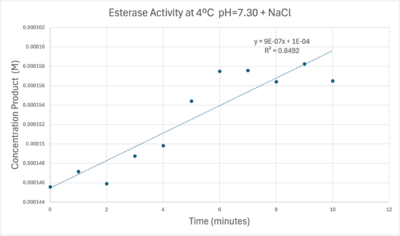
| - | [[Image: | + | [[Image:Concentrationcoldmolarabsorb12000ph7.png|400px|left|thumb|A trendline showing the relationship between time and product concentration from the data gathered from the PNPA+Protein solution with pH = 7, sodium chloride, and a cold environment.]] |
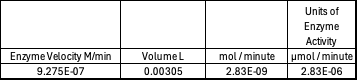
| - | + | Absorbance values can be transformed to units of concentration via the Beer-Lambert law. We must accept the approximation of the Molar Extinction Coefficient for PNPA hydrolysis at 428.7nm as 12000 M^-1 cm^-1. An example calculation is supplied in the table. Graphing time versus concentration and determining the slope of the line yields the enzyme's velocity in M/min. 1mg/mL of PNPA is saturating conditions which implies the Vmax is also the slope. The reaction volume total times Vmax yields Units of Enzyme Activity. This value can be used as a relative comparison tool for enzyme performance in given conditions. | |
| - | [[Image: | + | [[Image:Iceenzymeefficiencyscinotation.png|600px|left|thumb|Enzyme Activity at neutral pH at cold temperature with Chloride cofactor.]] |
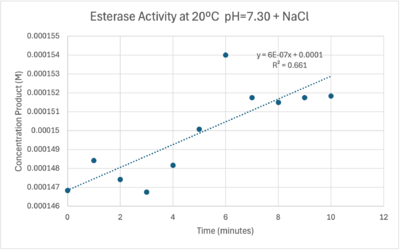
| - | + | The cold 4 degrees C reaction was changed to room temperature 20 degrees C. All other conditions remained constant to evaluate the effect of temperature on enzyme. Temperature increase negatively affects enzyme performance. Considering the cold loving nature of Chitinophaga pinensis, the enzyme being more active at a lower temperature is a reasonable conclusion. | |
| - | [[Image: | + | [[Image:Roomtempconcentrationtime.png|400px|right|thumb|Enzyme Activity at neutral pH at room temperature with Chloride cofactor.]] |
| - | + | [[Image:Sigfigroomtempenzymeactivity.png|600px|right|thumb|Enzyme Activity at neutral pH at room temperature with Chloride cofactor.]] | |
| - | [[Image: | + | [[Image:Enzyme units percentage increase.png|600px|left|thumb|4 degrees C yields 42.2% increase in Units of Enzyme Activity цmol/minute.]] |
| + | === Conclusion === | ||
| + | 42.2% increase in units of enzyme activity was a considerable gain. No replicates were done for this study. A future experiment would include replicates so t-test can be run to determine significance of the increase. Overall the temperature decrease had an effect on the conversion of PNPA to PNP. | ||
| - | + | A comparison model is the thermostable esterase from the moderate thermophile Bacillus circulans. Esterase activity was associated with a protein of molecular mass 95 kDa, composed of three identical subunits of 30 kDa. The esterase activity was thermostable with a maximum activity at 55 °C using PNPA assay. Literature reported units of enzyme activity in umol/minute at rate 790% greater than 4q7q. Taken as a comparison model, 4q7q rate is quite slow.<ref name="Thermo">Kademi, A.; Ait-Abdelkader, N.; Fakhreddine, L.; Baratti, J. Purification and characterization of a thermostable esterase from the moderate thermophile Bacillus circulans. Appl. Microbiol. Biotechnol., 2000, 54(1), 173-179, https://doi.org/10.1007/s002530000353</ref> | |
| + | The role of the chloride cofactor was not investigated thoroughly. Chloride ion was present in all reaction conditions, so it's independence was not tested. A future experiment would be testing the functionality of the enzyme without chloride in solution compared to with chloride ion. | ||
| + | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Current revision
Structural Analysis and Proposed Functionality of 4Q7Q
4Q7Q is a homodimeric protein complex that originates from the bacterial species Chitinophaga Pinensis and has a mass of 58.5 kDa. It is a member of the SGNH Hydrolase Superfamily with structural and sequential similarities to esterases and lipases. Current evidence suggests it causes the hydrolysis of esters and/or acetyl groups on lipids/lipid-like molecules via a catalytic triad-like active site.
| |||||||||||
References
- ↑ 1.0 1.1 4Q7Q. Protein Database, 2014. https://www.rcsb.org/structure/4Q7Q
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Nadzirin, N.; Gardiner, E.; Willett, P.; Artymiuk, P. J.; Firdaus-Raih, M. 2012. SPRITE and ASSAM: web servers for side chain 3D-motif searching in protein structures. Nucleic Acids Res., 40(Web Server Issue), W380-6.
- ↑ 3.0 3.1 Rio, T. G. D.; et al. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). Stand. Genomic. Sci., 2010, 2(1), 87-95. https://pmc.ncbi.nlm.nih.gov/articles/PMC3035255/
- ↑ 4.0 4.1 4.2 SGNH hydrolase superfamily. InterPro, 2017. https://www.ebi.ac.uk/interpro/entry/InterPro/IPR036514/
- ↑ 5.0 5.1 5.2 Rio, T. G. D.; et al. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). Stand. Genomic. Sci., 2010, 2(1), 87-95. https://pmc.ncbi.nlm.nih.gov/articles/PMC3035255/
- ↑ 6.0 6.1 6.2 6.3 Molgaard, A.; Kauppinen, S.; Larsen, S. Rhamnogalacturonan acetylesterase elucidates the structure and function of a new family of hydrolases. Struct., 2000, 8(4), 373-383. https://www.sciencedirect.com/science/article/pii/S0969212600001180?via%3Dihub
- ↑ UCSF Chimera--a visualization system for exploratory research and analysis. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004 Oct;25(13):1605-12.
- ↑ Akoh, C. C.; Lee, G.; Liaw, Y.; Huang, T.; Shaw, J. GDSL family of serine esterases/lipases. Prog. Lipid Res., 2004, 43(6), 534-552. https://pubmed.ncbi.nlm.nih.gov/15522763/
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 Holm L, Laiho A, Toronen P, Salgado M (2023) DALI shines a light on remote homologs: one hundred discoveries. Protein Science 23, e4519
- ↑ 10.0 10.1 10.2 10.3 Bugnon M, Röhrig UF, Goullieux M, Perez MAS, Daina A, Michielin O, Zoete V. SwissDock 2024: major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res. 2024, 52 (W1), W324-W332. DOI: 10.1093/nar/gkae300.
- ↑ 11.0 11.1 11.2 11.3 Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39 (Web Server issue), W270-W277. DOI: 10.1093/nar/gkr366
- ↑ 12.0 12.1 12.2 12.3 Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model., 2021, 61 (8), 3891–3898, DOI: 10.1021/acs.jcim.1c00203
- ↑ 13.0 13.1 13.2 13.3 Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem., 2010, 31 (2), 455–461, DOI: 10.1002/jcc.21334
- ↑ Miesfeld, R. L.; McEvoy, M. M. Biochemistry, 2nd ed.; W. W. Norton & Company, 2021
- ↑ Xia, L.; Qian, M.; Cheng, F.; Wang, Y.; Han, J.; Xu, Y.; Zhang, K.; Tian, J.; Jin, Y. The effect of lactic acid bacteria on lipid metabolism and flavor of fermented sausages. Food Biosci., 2023, 56, 103172.
- ↑ 16.0 16.1 Bornscheuer, U. T. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev., 2002, 26(1), 73-81. https://doi.org/10.1111/j.1574-6976.2002.tb00599.x
- ↑ Schober, I.; Koblitz, J.; Carbasse, J. S.; Ebeling, C.; Schmidt, M. L.; Podstawka, A.; Gupta, R.; Ilangovan, V.; Chamanara, J.; Overmann, J.; Reimer, L. C. BacDive in 2025: the core database for prokaryotic strain data. Nucleic Acids Res., 2024, 53(D1), D748-D756, https://doi.org/10.1093/nar/gkae959
- ↑ 18.0 18.1 Protein Activity Assays Student Module. BASIL. https://docs.google.com/document/d/1G-H0tskmSVRhC0qsDau-7-EzUcUYN8kBUHFZyBStYcs/edit?tab=t.0 (accessed 04/28/25).
- ↑ Kademi, A.; Ait-Abdelkader, N.; Fakhreddine, L.; Baratti, J. Purification and characterization of a thermostable esterase from the moderate thermophile Bacillus circulans. Appl. Microbiol. Biotechnol., 2000, 54(1), 173-179, https://doi.org/10.1007/s002530000353